RIP-seq Applications in Diseases: From Mechanism to Therapy
In the biology research system, the interaction network between RNA and protein, as the core link of gene expression regulation, is deeply involved in biological processes such as post-transcriptional regulation, cell fate determination and pathological process evolution. Traditional research methods have obvious limitations in analyzing the temporal and spatial specificity of RNA-protein dynamic interaction, quantifying binding affinity and identifying low-abundance interaction events.
The innovation of RNA Immunoprecipitation Sequencing (RIP-Seq) technology integrates the targeted enrichment ability of RNA immunoprecipitation with the advantages of high-throughput sequencing's complete transcriptome analysis, which provides a systematic solution for deciphering the RNA-protein interaction code. In recent years, this technology has made breakthroughs in the exploration of basic biological mechanisms, the discovery of disease markers and the verification of innovative drug targets.
Through several research cases, this paper shows the application and value of RIP-Seq technology in revealing the interaction mechanism between RNA and protein, exploring the molecular mechanism of diseases and tapping potential therapeutic targets.
Revealing Function Between RNA and Protein
Bladder cancer (BC) is the most common malignant tumor in urinary system, and the prognosis of patients with advanced or refractory BC is very poor. Epigenetic changes have been proved to play an important role in the pathogenesis of BC, among which N6- methyladenosine modification (m6A), as the most common RNA methylation modification, can participate in the occurrence and development of tumors, but its role and mechanism in BC are still unclear.
Insulin-like growth factor-2-mRNA-binding protein 1 (IGF-2bp1) belongs to a conservative family of RNA-binding carcinoembryonic proteins, which has a "carcinogenic" effect in tumor-derived cells by affecting the stability, translatability or localization of RNA. Studies have shown that IGF-2bp1 can preferentially recognize m6A-modified mRNA and promote its stability and translation. Some non-coding RNA may be involved in regulating the expression and function of IGF2BP1.
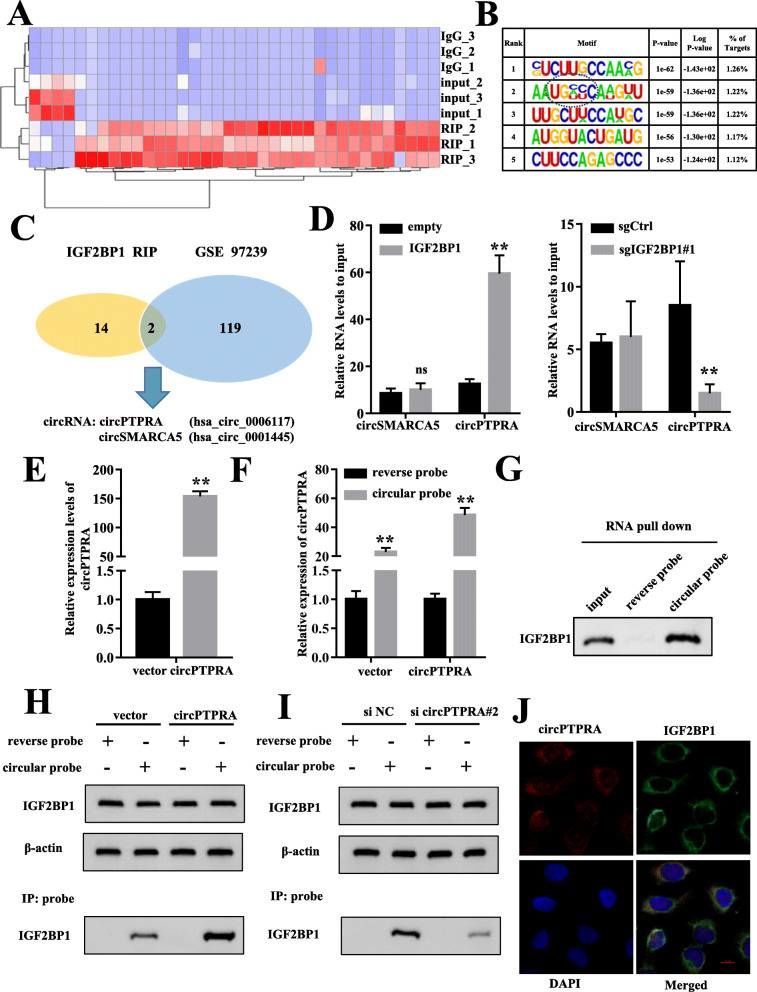 CircRNA interacting with IGF2BP1 detected by RIP-seq (Xie et al., 2021)
CircRNA interacting with IGF2BP1 detected by RIP-seq (Xie et al., 2021)
In order to explore the molecular mechanism of IGF-2bp1 in BC, RNA immunoprecipitation sequencing was performed to identify the RNA interacting with IGF-2bp1 in cells. In the enriched sequence, it was found that the binding site of IGF-2bp1 was rich in protein coding region (CDS) and typical m6A modified sequence characteristics, which was consistent with other previous studies.
At the same time, 16 candidate circRNA interacting with IGF2BP1 were identified. Next, three pairs of BC cancer tissues and corresponding non-cancer tissues were used for circRNA-seq, and it was found that circPTPRA (hsa_circ_0006117) and circSMARCA5 (hsa_circ_ 0001445) were significantly differentially expressed, which may interact with IGF-2bp1. The overexpression and knockout cell lines of IGF2BP1 confirmed the specific endogenous interaction between IGF2BP1 and circPTPRA. RNA-FISH and immunofluorescence analysis showed that circPTPRA was mainly located in cytoplasm and could bind to IGF2BP1.
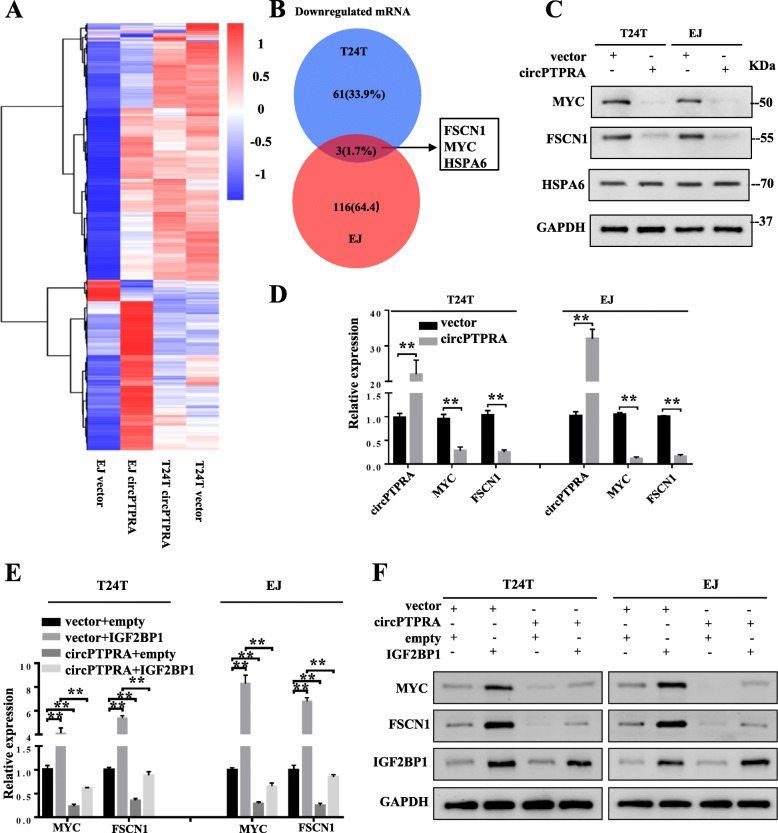 IGF2BP1 interacts with circPTPRA through its KH domain (Xie et al., 2021)
IGF2BP1 interacts with circPTPRA through its KH domain (Xie et al., 2021)
Service you may intersted in
Learn More:
Study on Carcinogenic Mechanism of CBNPs
With the acceleration of industrialization, the problem of environmental pollution is becoming more and more serious. In particular, carbon black nanoparticles (CBNPs), as the main component of air pollutants, have been classified as Class 2B carcinogens by the International Agency for Research on Cancer. Studies have shown that CBNPs can induce DNA damage in epithelial cells, but its potential and specific mechanism of carcinogenesis are not clear. Therefore, it is of great significance to study the relationship between DNA damage induced by CBNPs and carcinogenesis for understanding the carcinogenicity of environmental pollutants.
The researchers first established a model of malignant transformation of human bronchial epithelial cells through cell culture in vitro and CBNPs treatment. Then, the overexpression of MSH2 gene and circ_0025373 was identified by transcriptome sequencing and RIP-seq technology. Further, through a series of experiments, including RNA extraction and real-time quantitative PCR, the interaction between circ_0025373 and MSH2 and its influence on DNA damage were revealed.
First of all, 16HBE human bronchial epithelial cells exposed to carbon black nanoparticles (CBNPs) for a long time showed remarkable characteristics of malignant transformation in cell morphology and molecular level, in which the dynamic changes of epithelial-mesenchymal transition (EMT) markers and the continuous aggravation of DNA damage revealed the potential mechanism of cell carcinogenesis induced by CBNPs.
In the process of EMT, it was found by Western blot and qRT-PCR that with the increase of CBNPs generations (20 to 120 generations), the protein and mRNA expression levels of epithelial cell marker E-cadherin decreased gradually, while mesenchymal cell markers N-cadherin and Vimentin increased significantly. This phenomenon of "cadherin transformation" is a typical feature of EMT, which indicates that epithelial cells gradually lose the ability of intercellular adhesion and gain the migration and invasion characteristics of mesenchymal cells, laying the foundation for malignant transformation of cells.
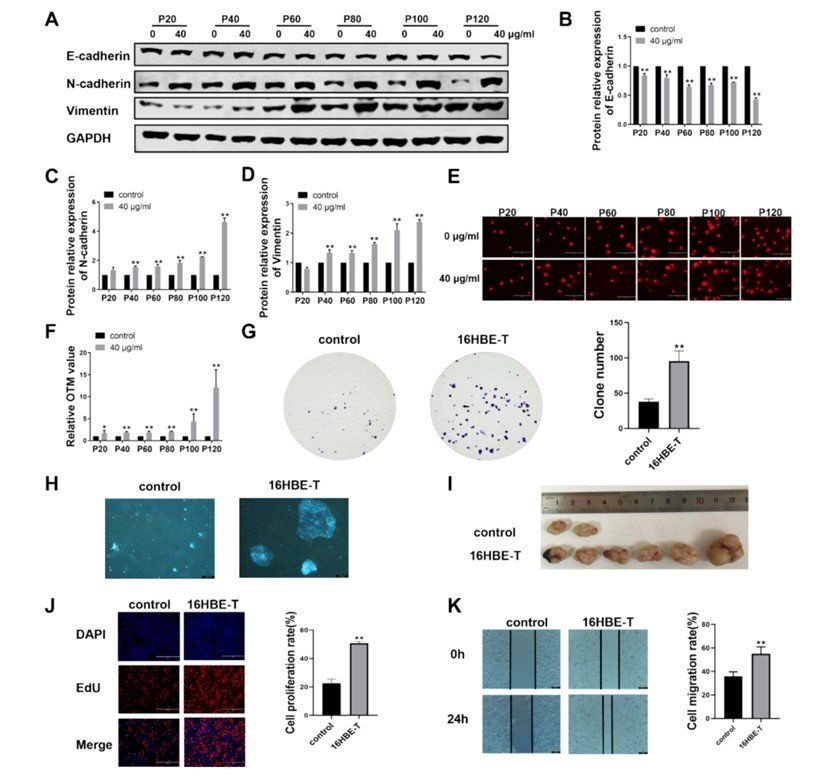 Malignant transformation of human bronchial epithelial cells induced by CBNPs exposure (Zhang et al., 2024)
Malignant transformation of human bronchial epithelial cells induced by CBNPs exposure (Zhang et al., 2024)
Subsequently, when exploring the molecular mechanism of malignant transformation of cells induced by CBNPs, the researchers found that MSH2, the core protein of DNA mismatch repair (MMR) pathway, and its related circular RNA (CIRC RNA)-CIRC _ 0025373 showed a significant upward trend in malignant transformation cells through transcriptome sequencing and RIP-seq, revealing that epigenetic regulation was involved in the carcinogenesis of CBNPs.
In order to explore the apparent regulatory factors related to MSH2, the researchers captured circRNA binding to MSH2 by RIP-seq technology, and found that circ_0025373 was highly expressed in 16HBE-T cells and lung cancer cells. QRT-PCR showed that the expression level of circ_0025373 was positively correlated with MSH2: in 16HBE cells exposed to CBNPs, the expression level of circ_0025373 gradually increased with the increase of generations, and its expression level in lung cancer cells was significantly higher than that in normal bronchial epithelial cells. The experiment of nuclear-cytoplasmic separation showed that circ_0025373 was mainly located in cytoplasm, which provided a spatial basis for its interaction with MSH2 protein in cytoplasm.
Mechanism study shows that circ_0025373 plays a regulatory role by directly binding to MSH2 protein. RNA pulldown assay confirmed that there was a physical interaction between circ_0025373 and MSH2 in 16HBE and 16HBE-T cells, while bioinformatics analysis showed that circ_0025373 sequence contained a specific motif that combined with MSH2. Further, the key areas of the combination of the two are determined by the prediction of catRAPI algorithm and the analysis of RIP-seq peak.
This combination not only affects the protein stability of MSH2, but also regulates its subcellular localization. In normal cells, MSH2 is mainly distributed in the nucleus to perform DNA repair function, while in cells exposed to CBNPs, the combination of circ_0025373 and MSH2 in the cytoplasm leads to the obstruction of its nuclear transport, thus weakening the ability of mismatch repair.
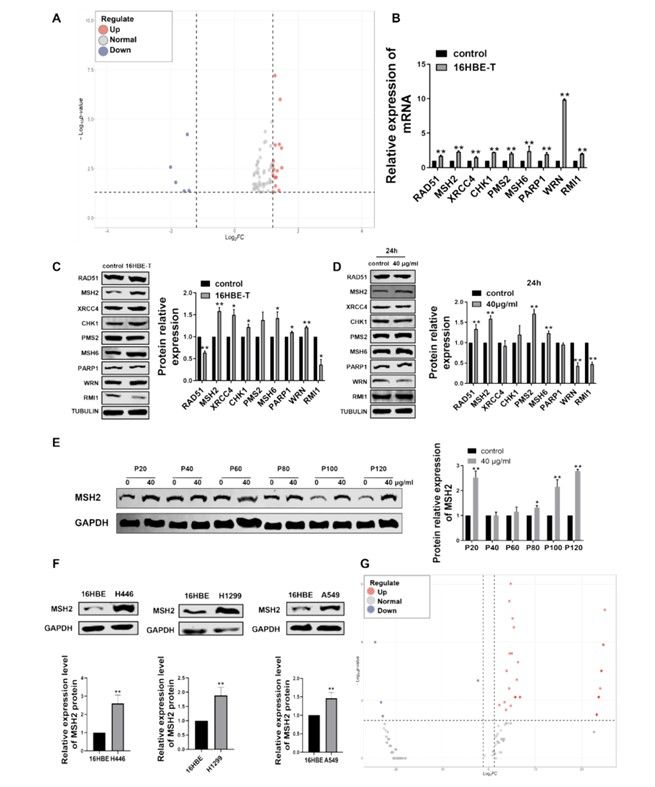 Screening and expression of MSH2 protein and circ_0025373 in malignant transformed cells induced by CBNP (Zhang et al., 2024)
Screening and expression of MSH2 protein and circ_0025373 in malignant transformed cells induced by CBNP (Zhang et al., 2024)
In addition, when exploring the interaction mechanism between circ_0025373 and MSH2 protein, the research team confirmed through multi-dimensional experiments that circ_0025373 regulates MSH2 through the triple mechanism of "binding-retention-function inhibition", that is, it directly binds MSH2 protein, stays in the cytoplasm, and prevents it from entering the nucleus to perform mismatch repair function, thus aggravating DNA damage and inhibiting malignant transformation of cells. This discovery not only reveals the dual role of circRNA in the response to DNA damage (early repair promotes vs. late repair inhibits), but also provides experimental basis for targeting circ_0025373-MSH2 axis to interfere with the carcinogenesis of nanoparticles, for example, blocking the combination of the two or regulating MSH2 nuclear transport through small molecular inhibitors, which may become a new strategy to prevent CBNPs-related lung cancer.
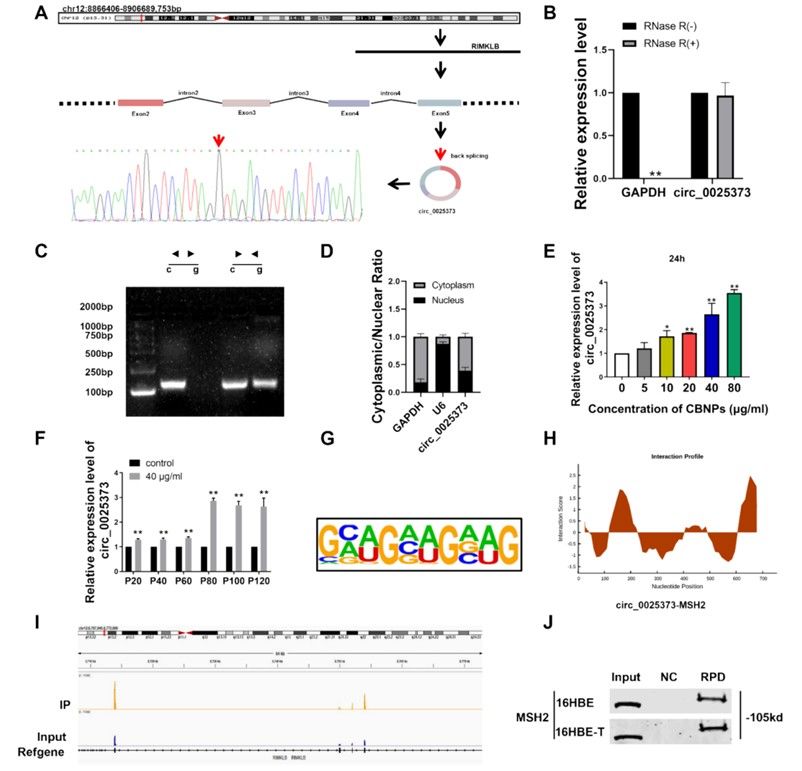 Circ_0025373 directly binds to MSH2 protein (Zhang et al., 2024)
Circ_0025373 directly binds to MSH2 protein (Zhang et al., 2024)
Explore Regulatory Mechanism of non-coding RNA
RIP-seq technology integrates the advantages of targeted enrichment and high-throughput sequencing, which can systematically capture ncRNA-protein interaction groups, accurately locate binding sites and quantify the interaction intensity.
Hypoxia Regulates mRNA Homeostasis
At first, the researchers compared the mRNA spectrum of AGO2 binding in HeLa cells under normoxic (21% O₂) and anoxic (1% O) conditions by RNA immunoprecipitation sequencing (RIP-seq). The results showed that after 24 hours of hypoxia treatment, the number of mRNA transcripts bound to AGO2 increased slightly (11,084 in normoxic group vs 13,176 in hypoxic group), but the cumulative score analysis showed that hypoxia significantly reduced the overall binding strength of mRNA to AGO2 (P = 2.272× 10, Mann-Whitney U test). Further analysis showed that the binding of 2100 mRNA transcripts to AGO2 was significantly weakened, but only 1528 were enhanced, suggesting that hypoxia may widely inhibit the capture efficiency of miRISC to target mRNA.
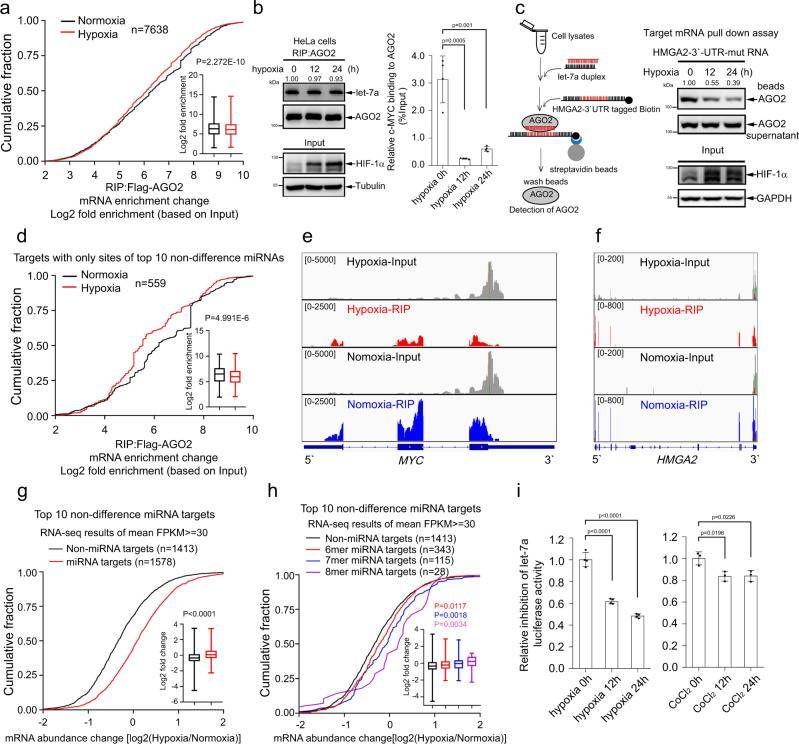 Hypoxia impairs miRNA-targeted mRNA loading to AGO2 and inhibits mRNA decay (Zhang et al., 2021)
Hypoxia impairs miRNA-targeted mRNA loading to AGO2 and inhibits mRNA decay (Zhang et al., 2021)
In order to eliminate the interference of miRNA expression profile changes, the research team found that hypoxia only affected a small part of miRNA production (such as pre-miR-192, pre-miR-31 and other long stem-loop precursors), while the expression of core mirnas (such as let-7a and miR-21) and the process of loading them into AGO2 were not significantly affected. This indicates that the inhibition of hypoxia on AGO2-binding mRNA is not due to the biological abnormality of miRNA, but directly affects the functional link of miRISC.
Combining RIP-seq and mRNA-seq data, the research team found that the M1-Ubi modification of AGO2 directly weakened its ability to recruit miRNA target mRNA. In the cells overexpressing LUBAC, the target mRNA of miRNA (such as c-MYC and HMGA2) bound by AGO2 decreased significantly, while knocking down HOIP reversed this phenomenon (Figure 6c-d, g-h). In mechanism, M1-Ubi modification interferes with the complementary pairing of AGO2 and mRNA through steric hindrance, which leads to miRISC's inability to effectively anchor the target.
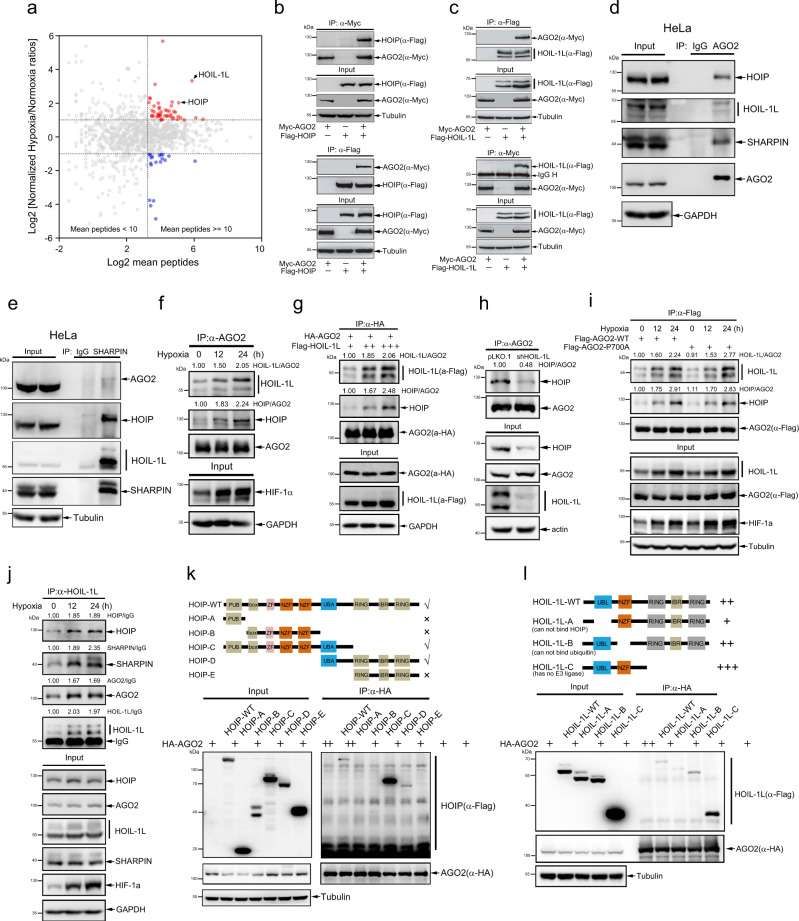 Hypoxia promotes the interaction of AGO2 with LUBAC (Zhang et al., 2021)
Hypoxia promotes the interaction of AGO2 with LUBAC (Zhang et al., 2021)
tRNAHis Guanylyltransferase Regulates Plant Heat Response
Through EMS mutation screening and geographical region phenotype analysis, the researchers identified a temperature-sensitive mutant aet1 in rice, which showed dwarfing, curly leaves and sterility in high temperature environment, but its phenotype was significantly relieved in low temperature environment. Map-based cloning showed that the single-base mutation (C→T, leading to amino acid substitution of Pro382Ser) of AET1 gene (encoding tRNAHis guanylate transferase) was the genetic basis of phenotype. The mutation significantly weakened the catalytic activity of AET1 protein-in vitro enzyme activity experiments showed that wild AET1 could catalyze GTP to be added to the 5' end of pre-tRNAHis, while mutant AET1P382S almost completely lost this function.
In order to explore the translation function of AET1, researchers identified 208 mRNA binding to AET1 at high temperature by RNA immunoprecipitation sequencing (RIP-seq), among which auxin response factor (OsARF) family genes were significantly enriched. Cluster thermogram showed that there were significant differences between wild-type and mutant genes such as OsARF19 and OsARF23 with upstream open reading frame (uORF). RIP-qPCR verification shows that AET1 not only binds to the main open reading frame (mORF) of OsARF gene, but also specifically binds to its uORF region, suggesting that it may be involved in translation regulation mediated by uORF.
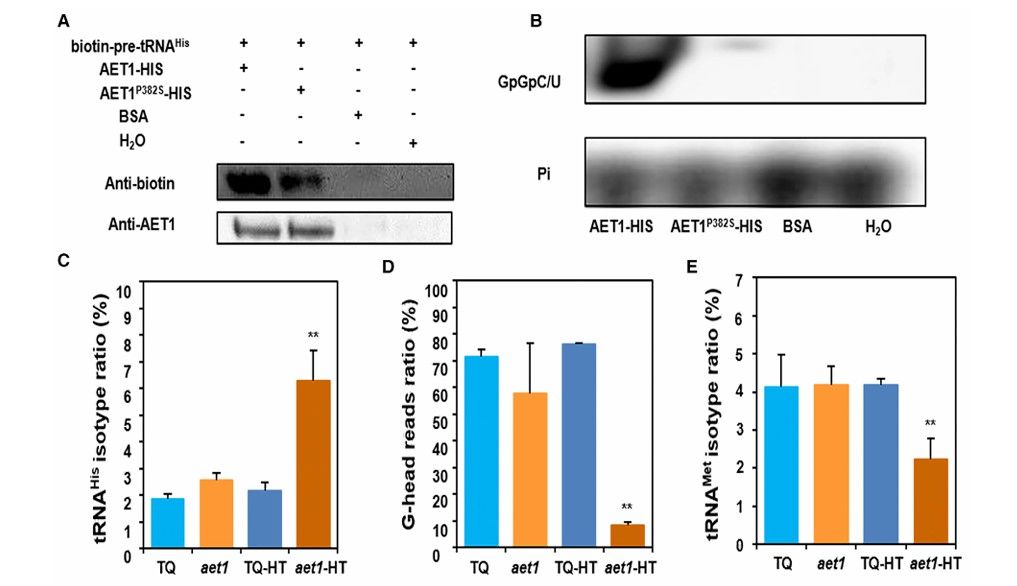 The aet1 mutation abolishes the guanylyltransferase activity of AET1 in vitro and disrupts the tRNA profile in vivo (Chen et al., 2019)
The aet1 mutation abolishes the guanylyltransferase activity of AET1 in vitro and disrupts the tRNA profile in vivo (Chen et al., 2019)
Drug Target Screening and Disease Treatment
Dilated cardiomyopathy (DCM) is one of the main causes of heart failure and heart transplantation. In recent decades, with the implementation of treatment strategies and early diagnosis, the prognosis of DCM patients has improved, but the treatment is still challenging. The potential causes of DCM are various, including gene mutation, virus infection, drug chemotherapy, autoimmune and systemic diseases.
The researchers detected the content of KSHV DNA and the expression level of kshv-miR-K12-1-5p in plasma of 696 patients with DCM, and followed up. The results showed that compared with the non-DCM group, the positive and quantitative titers of KSHV serum in DCM patients increased. During the follow-up period, DCM patients with positive KSHV DNA plasma are more likely to die due to cardiovascular reasons or heart transplantation. In heart tissue, compared with healthy donors, the content of KSHV DNA in DCM patients' hearts also increased.
Furthermore, 67 mRNAs related to Ago2 protein were identified by RIP-seq with Ago2 antibody. Through GO enrichment analysis, RNA pulldown, WB and fluorescence reporter gene experiments, several genes in the signal pathway of type I interferon, including OAS1, OAS2, OAS3, MX1, IFIT1, IFIT3, IRF7 and RSAD2, were identified as potential downstream targets of kshv-miR-K12-1-5p. It was also confirmed that kshv-miR-K12-1-5p directly targeted these genes, significantly reducing their expression, thus blocking the activation of type I interferon signal pathway activated by virus infection, and thus inhibiting its antiviral effect.
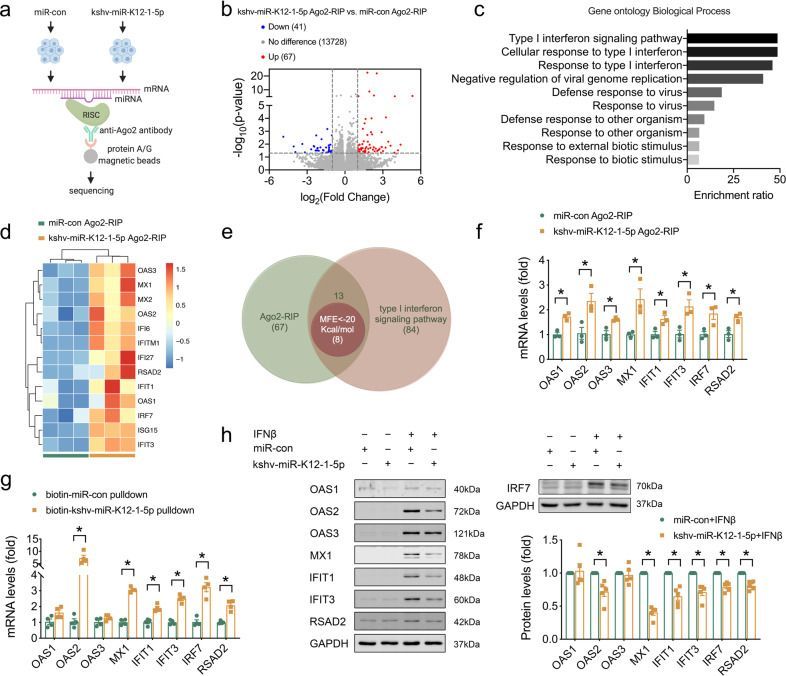 kshv-miR-K12-1-5p targeted lFN signaling pathway (Zhao et al., 2023)
kshv-miR-K12-1-5p targeted lFN signaling pathway (Zhao et al., 2023)
Conclusion
RIP-Seq as the core technology for analyzing RNA-protein interaction, shows multi-dimensional application value in biomedical research, and promotes a cross-level breakthrough from molecular mechanism to clinical transformation. This technology can systematically reveal the target and regulatory network of RBP by targeted capture of RNA molecules that bind to specific proteins and high-throughput sequencing, and provide key clues for analyzing complex biological processes.
References
- Xie F, Huang C., et al. "CircPTPRA blocks the recognition of RNA N6-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression." Mol Cancer. 2021 20(1): 68 https://doi.org/10.1186/s12943-021-01359-x
- Zhang H, Lu W., et al. "Circ_0025373 inhibits carbon black nanoparticles-induced malignant transformation of human bronchial epithelial cells by affecting DNA damage through binding to MSH2." Environ Int. 2024 191: 109001 https://doi.org/10.1016/j.envint.2024.109001
- Zhang H, Zhao X., et al. "Hypoxia regulates overall mRNA homeostasis by inducing Met1-linked linear ubiquitination of AGO2 in cancer cells." Nat Commun. 2021 12(1): 5416 https://doi.org/10.1038/s41467-021-25739-5
- Chen K, Guo T., et al. "Translational Regulation of Plant Response to High Temperature by a Dual-Function tRNAHis Guanylyltransferase in Rice. Mol Plant." 2019 2(8): 1123-1142 https://doi.org/10.1016/j.molp.2019.04.012
- Zhao Y, Li H., et al. "A Kaposi's sarcoma-associated herpes virus-encoded microRNA contributes to dilated cardiomyopathy." Signal Transduct Target Ther. 2023 8(1): 226 https://doi.org/10.1038/s41392-023-01434-3




