What Is DNA Sequencing?
DNA sequencing refers to the method employed to determine the order of the four nucleotide bases—adenine, thymine, cytosine, and guanine—that constitute a DNA molecule and convey critical genetic information. Within the DNA double helix, these four bases pair with specific partners to form units known as base pairs (bp): adenine (A) with thymine (T) and cytosine (C) with guanine (G). The human genome comprises approximately 3 billion such base pairs, providing the instructions for human creation and maintenance. The base-pairing structure facilitates the storage of copious genetic information, rendering DNA sequencing particularly suitable for this purpose. This complementary base pairing serves as the foundation for the mechanisms of DNA molecule duplication, transcription, and translation, and underlies the majority of DNA sequencing methods. Thanks to significant improvements in DNA sequencing technologies and methodologies, whole-genome sequencing has become both feasible and affordable. The objective of DNA sequencing is to determine the order of these bases, thereby enabling studies into gene function, genetic variation, evolutionary relationships, and other biological topics.
Service you may intersted in
DNA Sequencing Methods
DNA sequencing methodologies can typically be classified into two major categories: classical sequencing techniques and high-throughput sequencing approaches. A prime example of the classical approach is the Sanger sequencing method. First introduced in 1977 by British biochemist, Frederick Sanger and his colleagues, Sanger sequencing holds the distinction of being among the earliest DNA sequencing technologies. This technique draws on the principles of DNA polymerase and terminator nucleotides to interrupt the replication of the DNA chain, fragmenting the DNA molecule into sections of varying lengths. Following this, DNA polymerase and specific types of dNTPs (deoxyribonucleotide triphosphates) are employed to synthesize new DNA chains. Simultaneously, a unique marker is introduced to each type of base, permitting the determination of the DNA sequence through analysis of the sequence of these markers. For further information concerning this method, I would recommend you to refer to our tangible resource titled " Sanger Sequencing: Introduction, Principle, and Protocol".
Polymerase Chain Reaction (PCR) is a technique capable of amplifying specific DNA fragments in vitro. It executes cyclic processes such as the denaturation, annealing, and extension of DNA at distinctive temperatures, thereby generating numerous clones of the envisaged DNA fragment. Within the realm of DNA sequencing, PCR commonly serves to prepare the samples to be sequenced. An example of this is seen in Sanger sequencing, wherein a substantial quantity of DNA is typically required for the sequencing procedure. PCR facilitates the amplification of minute DNA sections, thereby lending feasibility to the sequencing experiment. Furthermore, within Sanger sequencing, PCR-amplified DNA fragments are often converted into templates viable for linear amplification, and sequencing is then carried out through chain termination reactions. Indeed, DNA sequences within PCR products can be decrypted in terms of their base sequence via the Sanger sequencing method.
The emerging next-generation sequencing (NGS), also known as massively parallel sequencing, has largely replaced Sanger sequencing due to its high throughput, cost-effectiveness, and rapidity. NGS can simultaneously determine the sequence of millions of fragments. It is characterized by short-read sequencing, requiring the construction of small fragment libraries, followed by deep sequencing, raw data preprocessing, DNA sequence alignment, assembly, annotation, and downstream analysis. The emerging third-generation sequencing, also known as long-read sequencing, including PacBio SMRT sequencing and Oxford Nanopore sequencing, allows the interrogation of billions of DNA and RNA templates while detecting variable methylation without bias. Long-read methods enable the detection of more variations, some of which cannot be observed solely through short-read sequencing.
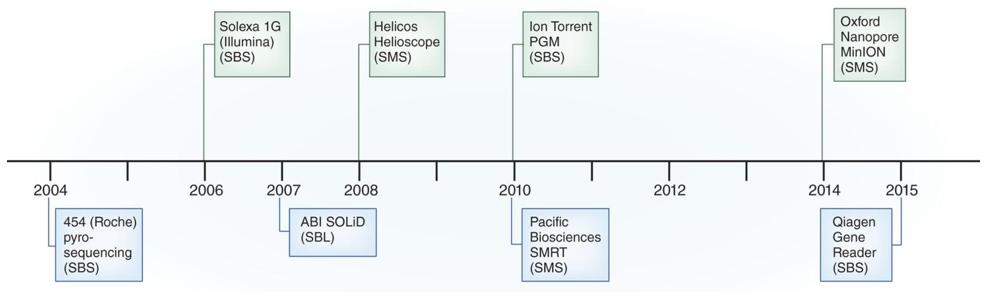
Figure 1. The history of DNA sequencing technologies.
Table 1. Sequencing Platform Comparison. (Dewey et al., 2012)
| Platform | Amplification | Sequencing | Detection | Read Length | |
| Second-generation sequencing platforms | 454 | Emulsion PCR on beads | Unlabeled nucleotide incorporation | Detection of light emitted by release of PPi | Variable (400 bp for single end sequencing) |
| SOLiD | Emulsion PCR on beads | Ligation of 2-base encoded fluorescent oligonucleotides | Fluorescence emission from labeled oligonucleotides | 75+35 bp | |
| Illumina | Array-based enzymatic amplification | Fluorescently labeled end-blocked nucleotide incorporation | Fluorescence emission from nucleotides | 2×100 bp | |
| Complete | Rolling-circle replication of short segments of DNA into nanoballs | Ligation of fluorescently labeled oligonucleotide probes | Fluorescence emission from oligonucleotide probes | 2×35 bp | |
| Third-generation sequencing platforms | Helicos | NA | Single dye-labeled nucleotides are added sequentially and incorporated by polymerases by use of single DNA molecular templates | Microscopy of fluorescently labeled nucleotides | 2×25–55 bp |
| Pacific Biosciences | NA | Incorporation of fluorescently labeled nucleotides by polymerases on solid support | Zero-mode waveguide imaging of fluorescent nucleotide incorporation by individual polymerases | 2×1000 bp | |
| Oxford nanopore | NA | Processive endo- or exonuclease activity feeds individual bases or whole DNA strands through protein or solid-state nanopores | Current disruption across nanopore corresponds to nucleotide structure | Variable | |
| Ion Torrent | Variable | DNA polymerase incorporation of unlabeled nucleotides added sequentially to solid-state microwells | Solid-state detection of hydrogen ions released by nucleotide incorporation | 200 bp |
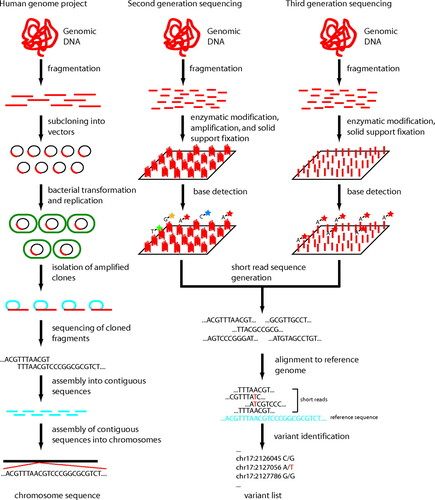
Figure 2. Three generations of human genome sequencing technology. (Dewey et al., 2012)
Applications of DNA Sequencing Technologies
DNA sequencing unveils the genetic information harbored within specific DNA segments, entire genomes, or complex microbial communities. Scientists harness this sequence data to decipher the genes and regulatory directives embedded within DNA molecules. Gene features such as open reading frames (ORFs) and CpG islands can be discerned through DNA sequence screening. Comparative analysis of homologous DNA sequences from different organisms facilitates evolutionary studies among species or populations. Importantly, DNA sequencing can elucidate genetic variations that may underlie diseases.
DNA sequencing is pivotal in various domains, encompassing medical applications such as disease diagnosis, treatment, and epidemiological studies. Sequencing bears the capacity to drastically reshape food safety and sustainable agriculture, including animal, plant, and public health arenas. It enables the enhancement of agriculture through efficacious animal and plant breeding, subsequently attenuating the risk of disease outbreaks. Moreover, DNA sequencing can be instrumental in safeguarding and ameliorating the natural environment of humans and wildlife.
In genomics research, DNA sequencing serves a critical role in deciphering an organism’s entire genomic sequence, facilitating our understanding of its genomic structure and composition. Transcriptomic research harnesses DNA sequencing to analyze the transcriptional activity of all genes within an organism under specific conditions. This enables researchers to discern patterns of gene expression and unravel underlying regulatory mechanisms. DNA sequencing is instrumental in proteomic research as well. By predicting protein-coding regions via DNA sequences, researchers can delve into studying the structure and functions of proteins.
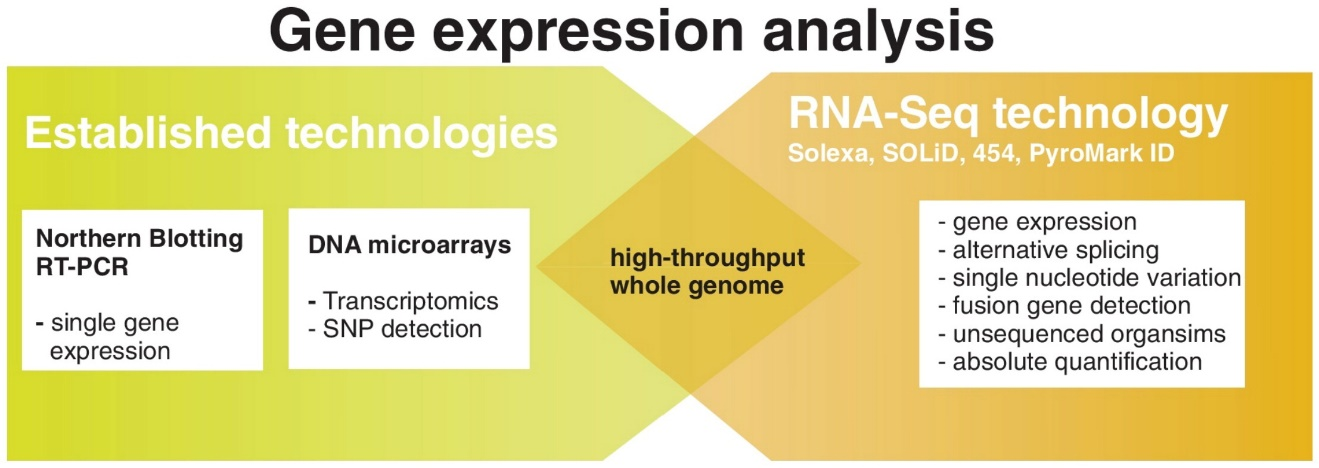
Figure 3. Transcriptome analysis using next-generation sequencing. (Mutz et al., 2013)
A significant application of DNA sequencing lies in disease diagnosis and treatment. It serves as a valuable tool in diagnosing genetic diseases, identifying tumorigenic gene mutations, detecting microbial infections amongst others, all of which contribute to the advancement of personalized medicine. In evolutionary biology, DNA sequences of different species are compared to infer evolutionary relationships and genetic variations. In forensic sciences, DNA sequence analysis plays a pivotal role in investigations and parentage testing. Additionally, the field of agriculture and food safety employs DNA sequencing for purposes including but not limited to crop variant modifications and to check for genetically modified components in food.
Daiger et al. demonstrate the efficacy of next-generation DNA sequencing in detecting disease-causing mutations, using Retinitis pigmentosa (RP), an inherited monogenic disease with significant genetic heterogeneity, as a test case. This study involved sequencing DNA from affected pairs in 21 families who exhibited autosomal dominant RP, while the sequencing targeted 1000 amplicons which spanned 249,267 unique bases of 46 candidate genes. Two different platforms were employed: the 454GS FLX Titanium (Roche Diagnostics) and GAIIx (Illumina/Solexa), with an average sequence depth of 70x and 125x, respectively. The analysis of over 9000 sequence variants led to the recognition of 112 as probable pathogenic candidates, which were further scrutinized with traditional di-deoxy capillary electrophoresis sequencing of additional family members and control subjects. The approach successfully identified five disease-causing mutations, accounting for 24% of the investigated families. The researchers conclude that next-generation sequencing is a valuable tool in detecting uncommon, novel mutations causing heterogeneous monogenic disorders such as RP, and its incorporation can aid in identifying disease-causing mutations in 65% of autosomal dominant RP cases.
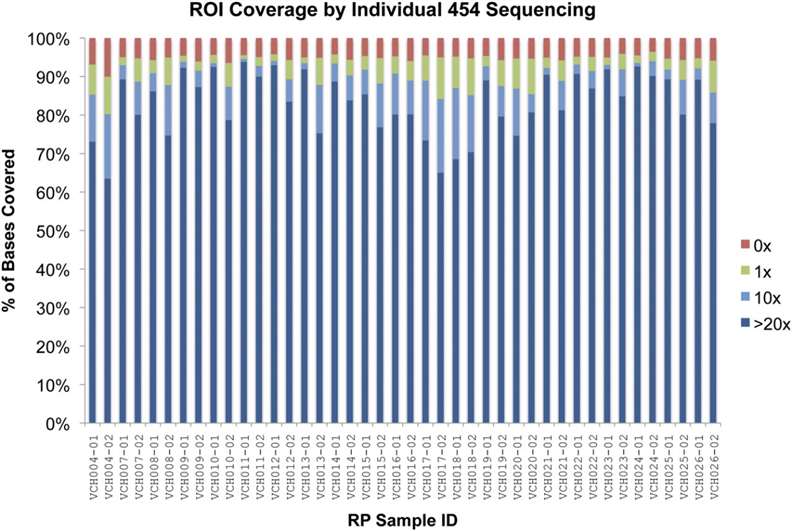
Figure 4. Coverage from 454GS FLX sequencing of individual samples (Roche Diagnostics, Indianapolis, IN). (Bowne et al., 2011)
The article by Daiger et al. details the application of a novel diagnostic sequencer tool for patients with retinitis pigmentosa (RP) – a highly genetic heterogeneous Mendelian disorder. They utilized a custom-made sequence capture array specifically designed to target the coding regions of known RP genes in DNA samples from five patients. After DNA enrichment and the creation of sequence libraries, high-throughput sequencing was undertaken either individually or in pools. Read alignment followed, identifying sequence variants mapped against the reference sequence. Pathogenicity was assessed through functional predictions and frequency in controls. The study successfully detected known homozygous PDE6B and complex heterozygous CRB1 mutations in two patients. Moreover, a novel homozygous missense mutation in the cyclic nucleotide-gated channel β1 (CNGB1) gene (c.2957A→T; p.N986I), predicted to be deleterious and absent in 720 control chromosomes, was identified in one patient where traditional genetic screening methods failed. The research validates the strength of high-throughput DNA sequencing coupled with DNA pooling as an effective diagnostic tool for genetic disorders with significant heterogeneity, such as RP. The pioneering technique increases the detection rate of disease-causing mutations and provides a promising pathway for future diagnosis and understanding of complex genetic diseases.
In essence, the continuous evolution and widespread application of DNA sequencing technology have equipped the arenas of life sciences with a formidable tool, catalyzing progress and development across numerous fields.
References:
- Mardis E R. DNA sequencing technologies: 2006–2016. Nature protocols, 2017, 12(2): 213.
- Dewey F E, Pan S, Wheeler M T, et al. DNA sequencing: clinical applications of new DNA sequencing technologies. Circulation, 2012, 125(7): 931-944.
- Shendure J, Aiden E L. The expanding scope of DNA sequencing. Nature biotechnology, 2012, 30(11): 1084-1094.
- DNA sequencing: methods and applications. BoD–Books on Demand, 2012.
- Shendure J, Balasubramanian S, Church G M, et al. DNA sequencing at 40: past, present and future. Nature, 2017, 550(7676): 345-353.
- Logsdon G A, Vollger M R, Eichler E E. Long-read human genome sequencing and its applications. Nature Reviews Genetics, 2020, 21(10): 597-614.
- Robledo D, Palaiokostas C, Bargelloni L, et al. Applications of genotyping by sequencing in aquaculture breeding and genetics. Reviews in aquaculture, 2018, 10(3): 670-682.
- Mutz K O, Heilkenbrinker A, Lönne M, et al. Transcriptome analysis using next-generation sequencing. Current opinion in biotechnology, 2013, 24(1): 22-30.
- Bowne S J, Sullivan L S, Koboldt D C, et al. Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing. Investigative ophthalmology & visual science, 2011, 52(1): 494-503.

Leave a Reply