DNA Methylation Research: An Overview of Method Selection, Technologies, and Research Frameworks
Introduction
DNA methylation is a pivotal epigenetic regulation mechanism facilitated by DNA methyltransferases (DNMTs). This process involves the covalent bonding of a methyl group at the carbon-5 position of cytosine, resulting in the formation of 5-methylcytosine (5mC). Alongside its derivatives like the "sixth base" 5-hydroxymethylcytosine and 5-formylcytosine, DNA methylation dynamically influences gene expression and demethylation through TET dioxygenases. This regulation plays a crucial role in embryonic development, tumor formation, aging, and is closely linked to conditions such as cancer and neurodegenerative diseases.
Over the past three decades, DNA methylation detection technologies have undergone significant transformation. Initially, these methods relied on chromatography to differentiate between methylated and unmethylated cytosine. Subsequent techniques involved indirect detection through methylation-sensitive restriction enzymes and 5mC antibody immunoprecipitation. A breakthrough in 1992 with bisulfite conversion technology enabled the chemical transformation to distinguish methylation sites, facilitating the first single-base resolution analysis.
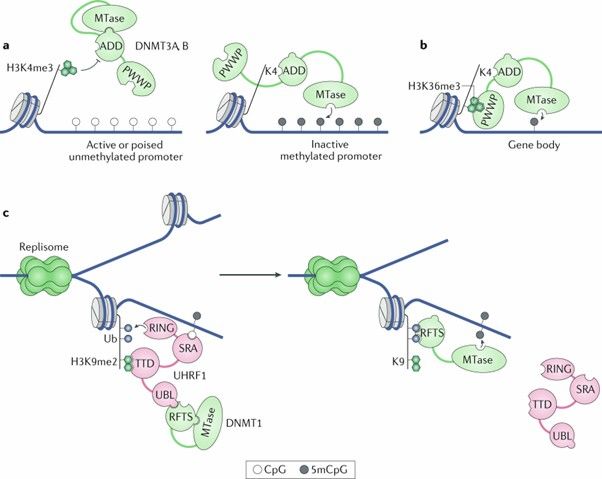 The diverse roles of DNA methylation in mammalian development and disease. (Greenberg, M.V.C., et al., 2019)
The diverse roles of DNA methylation in mammalian development and disease. (Greenberg, M.V.C., et al., 2019)
More recently, high-throughput technologies such as methylation microarrays (like the Infinium 850K) and third-generation sequencing (including nanopore sequencing) have accelerated the mapping of whole genome epigenomes. Meanwhile, targeted detection techniques such as pyrosequencing and digital PCR show potential for early cancer screening clinical applications.
Yet, significant differences exist among these methods. Whole-genome bisulfite sequencing (WGBS), though considered the "gold standard," faces challenges like DNA degradation and high costs. Methylation microarrays offer broad compatibility, supporting FFPE samples and are preferred for large cohort studies. Emerging enzyme-based methods (EM-seq) and single-cell sequencing techniques (scWGBS) address issues related to small sample quantities and cellular heterogeneity.
This article systematically explores the biological significance of DNA methylation and its detection methodologies, covering their technical principles, application scenarios, and research strategies. It aims to provide a methodological reference for basic research, focusing on the necessity, technical comparisons, experimental design, and case studies to aid readers in selecting suitable research tools.
Functionality of DNA Methylation Modifications
DNA methylation operates through three primary regulatory models:
1. Classical Model of DNA Methylation Regulation: In this traditional framework, methylated CpG island promoters attract transcription-repressive MBD proteins, preventing transcription factor binding. Conversely, unmethylated CpG islands are accessible to transcription factors.
2. New Model of DNA Methylation for Transcription Regulation: Genes with methylated CpG island promoters are inhibited by repressive complexes containing MBDs. Additionally, enhancer methylation can block transcription factor attachment. Active genes featuring unmethylated CpG island promoters often associate with activator complexes that include a CXXC domain. Transcription factors may also bind to non-methylated enhancers. Furthermore, highly methylated gene bodies of active genes aid in suppressing inadvertent transcription.
3. Decoupling of DNA Methylation from Transcription Initiation Inhibition: In certain scenarios, promoters with low-density CpG methylation undergo active transcription. Repressive MBD proteins do not engage with these promoters, though the underlying reasons remain unclear. Additionally, sequences with low-density CpG, including enhancers and promoters, may facilitate binding by activating transcription factors. The H3K4me3 histone mark is an example associated with active transcription processes.
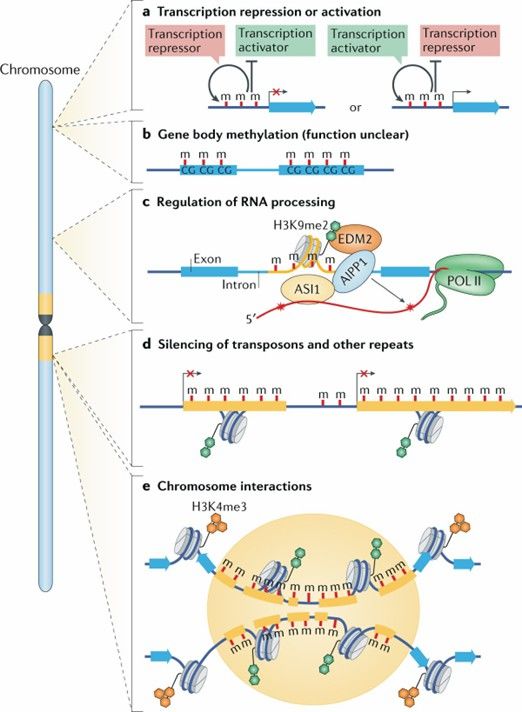 Dynamics and function of DNA methylation in plants. (Zhang, H., et al., 2018)
Dynamics and function of DNA methylation in plants. (Zhang, H., et al., 2018)
How much DNA methylation occurs
To effectively analyze human DNA methylation, it's essential to understand the fundamental aspects of methylation within the human genome. Humans possess approximately 3 billion base pairs, and DNA methylation predominantly occurs at CpG dinucleotides. The genome contains roughly 28 million CpG sites, with 60-80% of these sites typically methylated. CpG islands, which are CpG-rich sequences often found in promoter regions, are usually unmethylated. However, in tumor cells, the overall level of methylation decreases to 20-50% compared to normal cells. This change implies that the methylation status of about 2.8 to 16.8 million CpG sites is altered. In essence, while global methylation levels drop in tumor cells, there's a notable increase in methylation levels at the CpG sites within gene promoter regions.
Recommended Reading on DNA Methylation
For foundational insights into human and mouse basic medical research and cellular biology, consider the following reviews:
- "DNA methylation in mammalian development and disease." Nat Rev Genet. 2024
- "DNA methylation: old dog, new tricks?" Nat Struct Mol Biol. 2014, 21(11): 949
For insights into plant biology:
- "Epigenetic gene regulation in plants and its potential applications in crop improvement." Nat Rev Mol Cell Biol. 2024
- "Non-canonical RNA-directed DNA methylation." Nat Plants. 2016, 2(11): 16163
Service you may intersted in
Learn More:
Why Detect DNA Methylation
Before we delve into the reasons for detecting DNA methylation, it's important to ask: what questions can DNA methylation profiling answer? DNA methylation is well-known for its ability to regulate gene expression. Thus, analyzing DNA methylation can provide valuable insights into the regulatory mechanisms governing gene expression. DNA methylation plays a role in a variety of crucial biological processes, including early embryonic development, genomic imprinting, X-chromosome inactivation, silencing of repetitive sequences, and the development and metastasis of cancer. Furthermore, DNA methylation serves as a significant biomarker for tumors.
1. Uncovering the "Hidden Rules" of Gene Expression
DNA methylation finely tunes cell functions by repressing or activating gene transcription:
- Gene Silencing: High methylation levels in promoter regions (e.g., in tumor suppressor genes) prevent transcription factor binding, leading to gene silencing.
- Gene Activation: Low methylation in gene bodies may enhance transcription elongation, supporting cell-specific functions, such as the expression of synaptic proteins in neurons. This "on-off" mechanism explains why cells with identical DNA sequences, like skin and liver cells, perform vastly different tasks.
2. Key Clues to Life's Mysteries
DNA methylation is integral to numerous core biological processes:
- Embryonic Development: After fertilization, genomes undergo widespread methylation erasure and reconstruction to ensure stem cell pluripotency. For example, abnormal methylation of the imprinted gene H19 can lead to developmental abnormalities in the fetus.
- Genomic Imprinting: Selective expression of maternal or paternal genes (like in Prader-Willi syndrome) relies on methylation marks.
- X-Chromosome Inactivation: In female cells, the silencing of one X chromosome is driven by the methylation of the XIST gene promoter.
- Cancer Metastasis: Tumor cells acquire invasive capabilities through methylation reprogramming of metastasis-related genes, such as TWIST1.
3. The "Molecular Probe" for Cancer Early Screening and Diagnosis
DNA methylation markers are ideal targets for tumor detection due to their high stability and tissue specificity:
- Non-invasive Screening: The detection of Septin9 gene methylation in the blood of colorectal cancer patients is clinically applied, offering early warning without the need for colonoscopy.
- Precise Typing: The methylation level of CDO1 in lung cancer can differentiate between adenocarcinoma and squamous cell carcinoma, guiding treatment decisions.
- Efficacy Monitoring: In breast cancer patients, dynamic changes in the methylation of circulating tumor DNA (ctDNA) post-treatment can provide real-time insights into drug resistance.
Summary of DNA Methylation Detection Methods
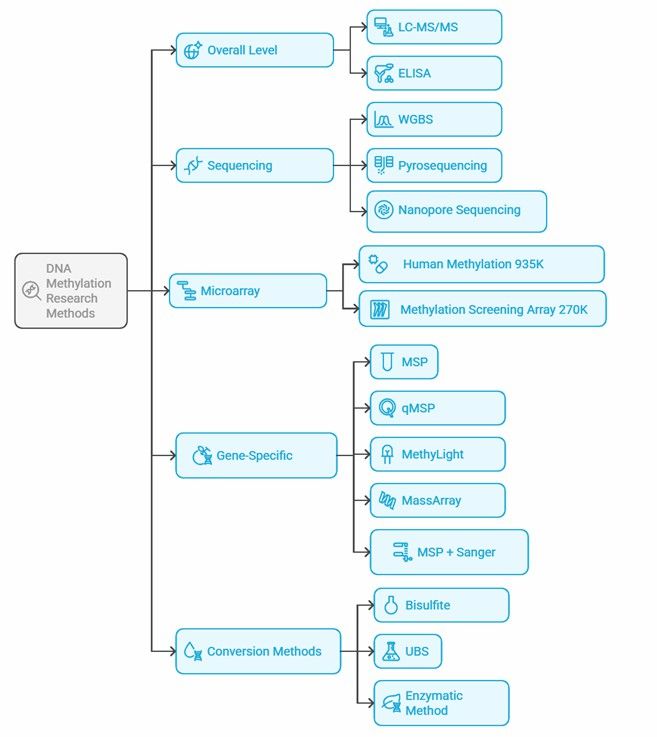 Mind map of DNA methylation research methods.
Mind map of DNA methylation research methods.
DNA methylation detection methods vary widely, and the choice depends on the specific research objectives, sample types, and cost considerations. Here's a concise overview of the primary methods:
1. Global Level Detection:
- LC-MS/MS: Highly accurate and sensitive, considered the "gold standard," but costly and complex to operate.
- ELISA: Fast and cost-effective but prone to interference and lower sensitivity.
2. Sequencing Technologies:
- WGBS: Provides single-base resolution, ideal for comprehensive studies, though complex and expensive.
- Pyrosequencing: Quantifies methylation in short fragments, suitable for quantitative analysis but with operational complexities.
- Nanopore Sequencing: Directly detects methylation without chemical conversion, promising but still developing and costly.
3. Microarray Technology:
- Used for large-scale, targeted analysis, more affordable than WGBS, and commonly used in clinical and cohort studies.
4. Specific Gene Detection:
- MSP: Simple and inexpensive, ideal for specific gene sites but involves complex primer design.
- MethyLight: Offers high-precision quantification at specific sites using probe-based methods.
- Digital PCR: More sensitive than MethyLight, effective for low methylation levels.
- HRM Analysis: Fast detection of methylation differences, requires precise temperature control.
- PCR + Sanger Sequencing: Confirms methylation sites with high accuracy, the "gold standard."
- MassArray (MALDI-TOF): High throughput, cost-effective, differentiates between methylated and unmethylated fragments after bisulfite conversion.
Detailed Overview of DNA Methylation Detection Methods
This section provides an overview of various DNA methylation detection methods, including techniques for assessing global methylation levels, sequencing-based approaches, microarray technologies, and gene-specific detection methods.
Global Methylation Level Detection
To assess methylation levels across the entire genome, the following methods can be employed:
1. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS):
- This method digests genomic DNA into single nucleotides (such as A, T, G, C, 5mC, and 5hmC) and uses mass spectrometry in Multiple Reaction Monitoring (MRM) mode to measure each base's content. This allows calculation of DNA methylation levels (5mC%) and simultaneous detection of 5hmC. Despite its extremely high accuracy and sensitivity, making it the gold standard for detecting whole-genome 5mC content, LC-MS/MS is expensive and complex to operate, thus unsuitable for high-throughput studies.
2. ELISA Methylation Detection:
- This method utilizes antibodies to detect 5mC levels and, via a standard curve, calculate DNA methylation levels (5mC%). ELISA is simple to use, fast, and available as commercial kits, but its reliance on antibody specificity may limit sensitivity and accuracy, especially in samples with subtle methylation changes.
Sequencing-Based Methylation Detection
To obtain site-specific and sequence-level information on DNA methylation, sequencing methods are ideal, some DNA methylation sequencing methods are described below:
1. Whole Genome Bisulfite Sequencing:
- WGBS combines bisulfite conversion with high-throughput sequencing to detect DNA methylation at single-base resolution across the entire genome. It covers all methylation sites and can be coupled with targeted techniques to detect low-frequency methylation signals, avoiding issues with repetitive sequences and SNPs. As sequencing costs decline and technology advances, WGBS will become more prevalent.
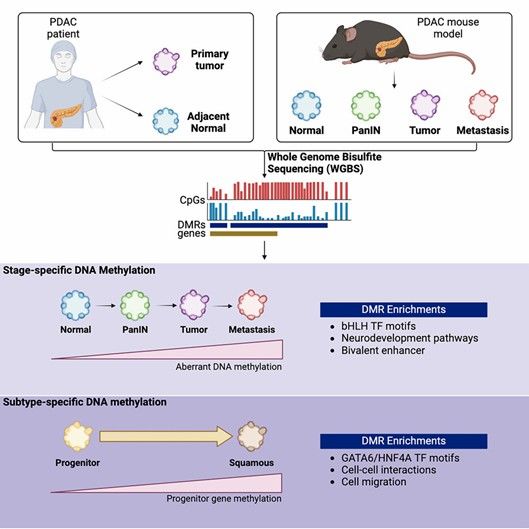 Whole-genome bisulfite sequencing identifies stage- and subtype-specific DNA methylation signatures in pancreatic cancer. (Wang, Sarah S., et al., 2024)
Whole-genome bisulfite sequencing identifies stage- and subtype-specific DNA methylation signatures in pancreatic cancer. (Wang, Sarah S., et al., 2024)
2. Oxidative Bisulfite Sequencing (oxBS-seq):
- By combining traditional BS-seq with chemical oxidation using potassium ruthenate (KRuO₄), this method oxidizes 5hmC to 5fC, which, along with unmodified C, converts to U upon bisulfite treatment, while 5mC remains unchanged. The combined use of oxBS-seq and BS-seq can also achieve single-base resolution detection of 5hmC.
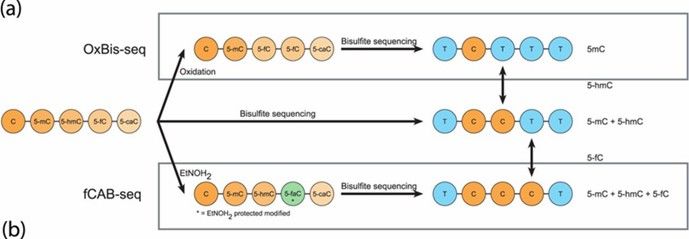 Principal scheme of oxBS-seq and fCAB-seq methods. (Becker, Daniel, et al., 2014)
Principal scheme of oxBS-seq and fCAB-seq methods. (Becker, Daniel, et al., 2014)
3. Reduced Representation Bisulfite Sequencing (RRBS):
- RRBS employs restriction enzymes (like MspI) to selectively cut the genome, enriching promoter regions and CpG islands for bisulfite sequencing. This approach increases sequencing depth in high-CpG regions, reducing costs and making it suitable for large-scale clinical research.
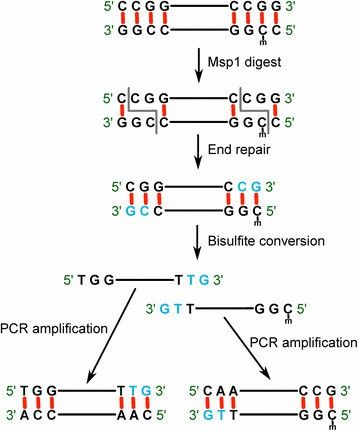 RRBS concept and workflow. (Baheti, Saurabh, et al., 2016)
RRBS concept and workflow. (Baheti, Saurabh, et al., 2016)
4. Single-Cell Whole Genome Methylation Sequencing (scWGBS):
- With challenges in library construction for single-cell DNA methylation, new scWGBS technology utilizes linear amplification and single-tube library construction to reduce bias, enabling high-precision methylation analysis for rare samples.
5. Amplicon Methylation Sequencing:
- Designed for specific gene regions using methylation-specific primers, this method employs PCR amplification of bisulfite or oxidized treated DNA, followed by high-throughput sequencing to analyze methylation status, suitable for precise target gene methylation analysis.
6. Hydroxymethylated DNA Immunoprecipitation Sequencing (hMeDIP-seq):
- Using antibodies specific to 5hmC, this method enriches hydroxymethylated DNA fragments for high-throughput sequencing, suitable for studying the role of 5hmC in gene regulation, disease, and embryonic development.
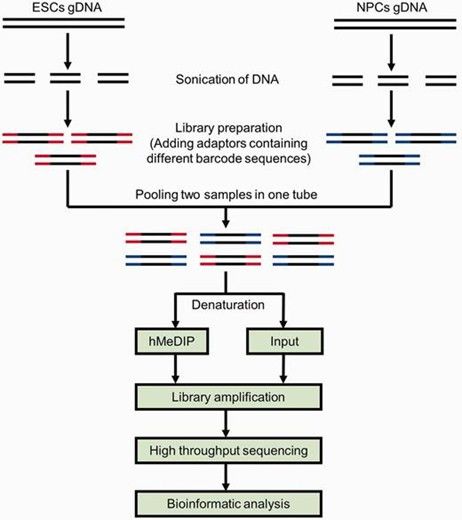 Schematic diagram of the comparative hMeDIP-seq method. (Tan, Li, et al., 2013)
Schematic diagram of the comparative hMeDIP-seq method. (Tan, Li, et al., 2013)
7. Enzymatic Methylation Sequencing (EM-seq):
- EM-seq overcomes WGBS limitations of DNA degradation from bisulfite by using TET2 enzymes and oxidation enhancers to convert 5mC and 5hmC to 5caC, then processing with APOBEC deaminase for methylation detection. This protects DNA integrity and enhances data quality.
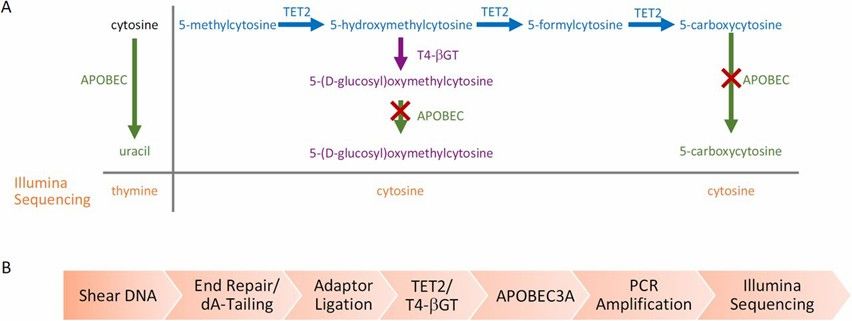 Enzymatic Methyl-seq mechanism of action and workflow. (Vaisvila, Romualdas, et al., 2021)
Enzymatic Methyl-seq mechanism of action and workflow. (Vaisvila, Romualdas, et al., 2021)
8. Pyrosequencing:
- Combining bisulfite treatment with pyrosequencing, this method detects methylation in specific target regions by comparing C and T ratios at individual sites. Fast and straightforward, pyrosequencing is ideal for short fragment analysis (20-50bp) but struggles with long homopolymer regions.
9. TET-Assisted Pyridine Borane Sequencing (TAPS):
- TAPS eschews bisulfite treatment by using TET1 oxidase to oxidize 5mC and 5hmC into 5caC, which is then reduced by pyridine borane to DHU, ultimately converting C to T during PCR. TAPS is non-destructive, reducing DNA loss and improving data quality at lower costs compared to traditional WGBS.
10. Nanopore Sequencing:
- This technique directly distinguishes methylated from unmethylated bases via changes in electric signals as DNA passes through nanopores, avoiding bisulfite treatment. Offering simultaneous methylation and sequence data, nanopore sequencing supports long-read sequencing. However, it remains costly, with ongoing improvements needed in data analysis.
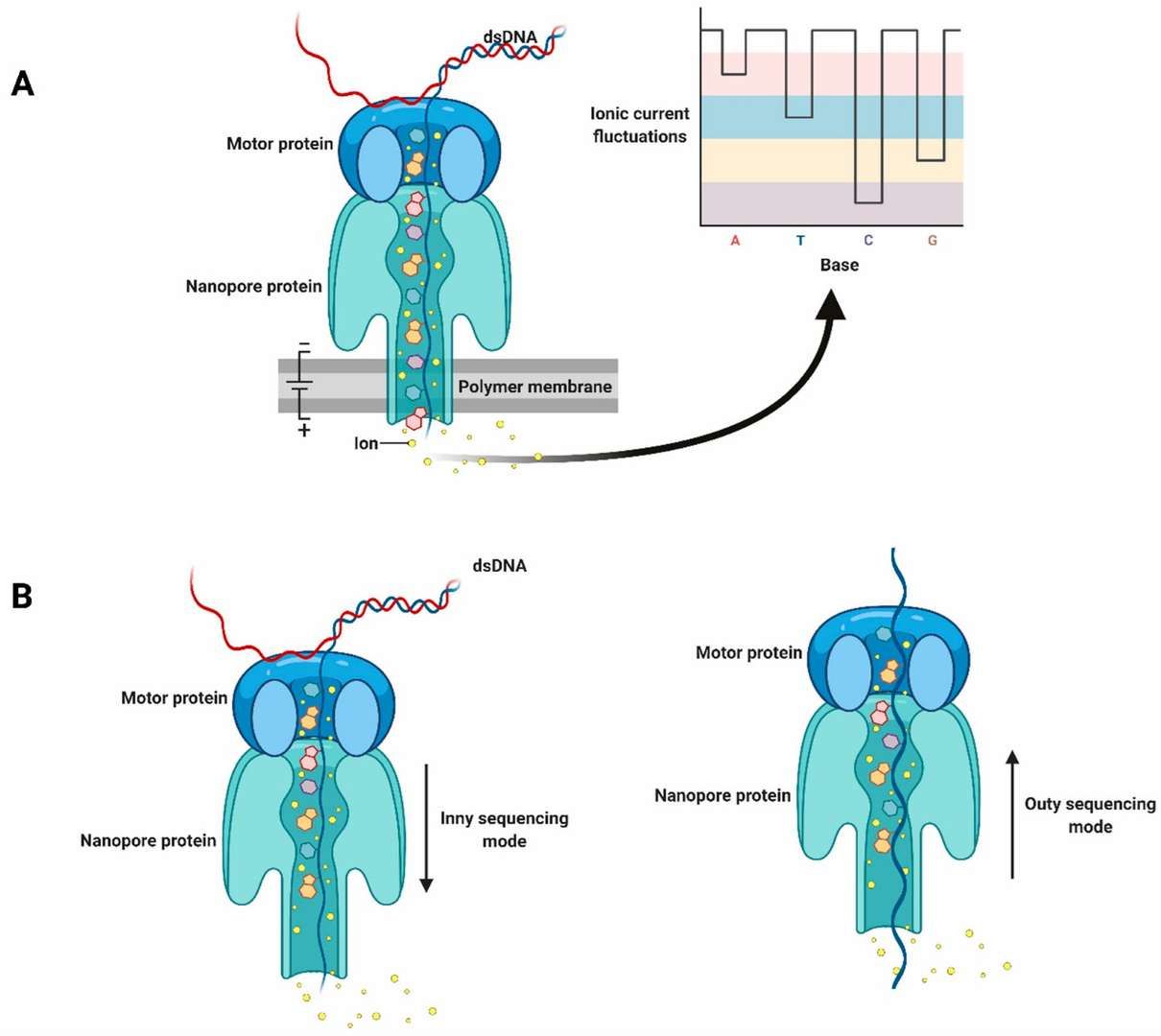 Schematic diagram of nanopore sequencing technology. (Javaran, Vahid Jalali, et al., 2021)
Schematic diagram of nanopore sequencing technology. (Javaran, Vahid Jalali, et al., 2021)
Service you may intersted in
Learn More:
DNA Methylation Microarrays
In large-scale cohort studies of whole-genome DNA methylation, the most feasible approach is often microarray technology rather than WGBS, primarily due to the high costs associated with sequencing and data processing. DNA methylation microarrays leverage hybridization of bisulfite-treated DNA probes, targeting CpG-rich cytosines of interest (both methylated and unmethylated).
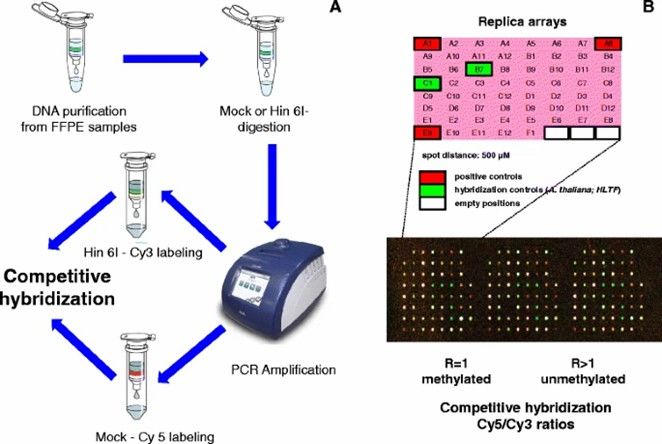 Microarray-mediated methylation assay. (Li, Y., Melnikov, A.A., et al., 2015)
Microarray-mediated methylation assay. (Li, Y., Melnikov, A.A., et al., 2015)
The 450K array, for example, involves the bisulfite conversion of 0.5-1μg of genomic DNA. Converted DNA hybridizes with an array of pre-designed methylation-specific probes: one set targets methylated cytosines, and another targets unmethylated cytosines. The probe's single base at the 3' CpG site incorporates a labeled and fluorescently tagged nucleotide (ddNTP). An Illumina iScan scanner then scans the bead array, detecting the fluorescence signal ratio.
The DNA methylation proportion at each CpG site is quantified as a β value, calculated using the following formula:
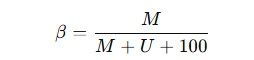
Where ( M ) is the intensity of the methylated signal, ( U ) is the intensity of the unmethylated signal, and 100 is a constant offset for adjustment when signal intensities are low. The β value ranges from 0 (0% methylation) to 1 (100% methylation).
DNA methylation microarrays are vital tools for studying DNA methylation. Compared to whole-genome methylation sequencing, methylation microarrays offer advantages such as lower starting material requirements, shorter processing times, and flexibility in sample types. They can be applied to FFPE (formalin-fixed paraffin-embedded) samples and valuable clinical specimens, thus playing a significant role in methylation detection technology.
To date, the Infinium Human Methylation 450K and Methylation EPIC v1.0 (850K) arrays have been widely adopted by researchers to gather epigenomic data, with increasing numbers of related publications each year. In 2023, Illumina launched the 935K array, covering over 935,000 marker sites. The 935K array retains compatibility with the 850K while removing 107,000 non-functional probes caused by SNPs, cross-hybridization, and multi-mapping, and adding at least 186,000 new probes, providing a more robust tool for clinical DNA methylation research.
Additionally, the Infinium Methylation Screening Array 270K is another DNA methylation chip from Illumina, covering approximately 270,000 CpG sites. Designed for rapid screening, it enables the analysis of DNA methylation in large clinical sample sets, suitable for preliminary methylation screening and validation. Although it has lower resolution, it remains effective for initial studies and high-throughput analysis.
It's important to note that DNA methylation microarrays are predominantly used for human samples. When dealing with large-scale clinical samples and well-defined research objectives, DNA methylation microarray technology is the most suitable method.
Service you may intersted in
Gene-Specific Methylation Detection Methods
Several methods are specifically designed to assess DNA methylation at gene-specific sites. Each method has unique features and advantages suitable for different research needs:
(1) Methylation-Specific PCR (MSP):
MSP is a straightforward and rapid method for qualitatively assessing DNA methylation. It can quickly determine the methylation status of CpG sites within CpG islands. The key is to design two pairs of primers for specific regions using tools like Methprimer for both methylated and unmethylated sequences. MSP's simplicity and speed are major advantages. For relative quantification, qMSP (Quantitative Methylation-Specific PCR) uses fluorescent dyes to indicate methylation levels by measuring fluorescence and Ct values.
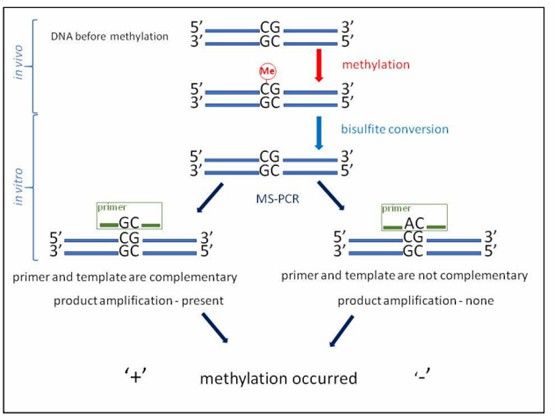 Reaction scheme for MS-PCR (DNA methylation has occurred). (Gryzinska, Magdalena, et al. 2019)
Reaction scheme for MS-PCR (DNA methylation has occurred). (Gryzinska, Magdalena, et al. 2019)
(2) MethyLight:
This method uses fluorescent probes to detect methylation. Methylation-specific primers are labeled with FAM, while unmethylated-specific primers are labeled with VIC. During real-time PCR, fluorescence signals are detected to determine methylation status. By measuring the Ct value, methylation can be quantitatively analyzed. MethyLight typically requires less than 100ng of bisulfite-converted DNA per PCR reaction and utilizes standards for quantification.
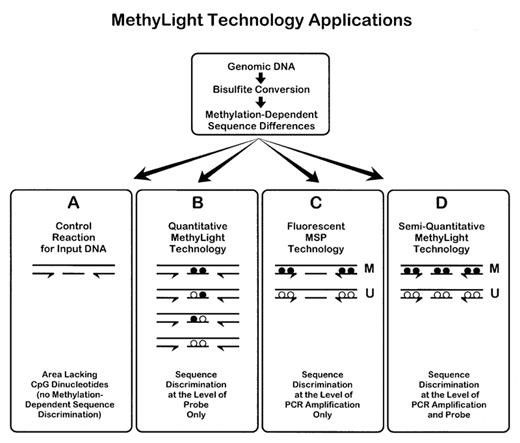 Schematic of the theoretical basis of MethyLight technology. (Eads, Cindy A., et al. 2000)
Schematic of the theoretical basis of MethyLight technology. (Eads, Cindy A., et al. 2000)
(3) Digital PCR (dPCR):
Digital PCR provides absolute quantification based on "single molecule counting," making it ideal for low-level methylation detection. Its key benefits include:
- High Sensitivity: Direct detection of methylation at the single-molecule level, suitable for low-concentration samples or rare methylation sites.
- Direct Quantification: Compared to MethyLight, it quantifies DNA copies directly without standard curves, with minimal impact from PCR efficiency.
- Strong Anti-Interference: Distributes sample uniformly across numerous micro-reaction units, diluting PCR inhibitors and minimizing interference.
dPCR is thus suitable for complex samples like paraffin-embedded tissues or circulating cell-free DNA (cfDNA) in plasma.
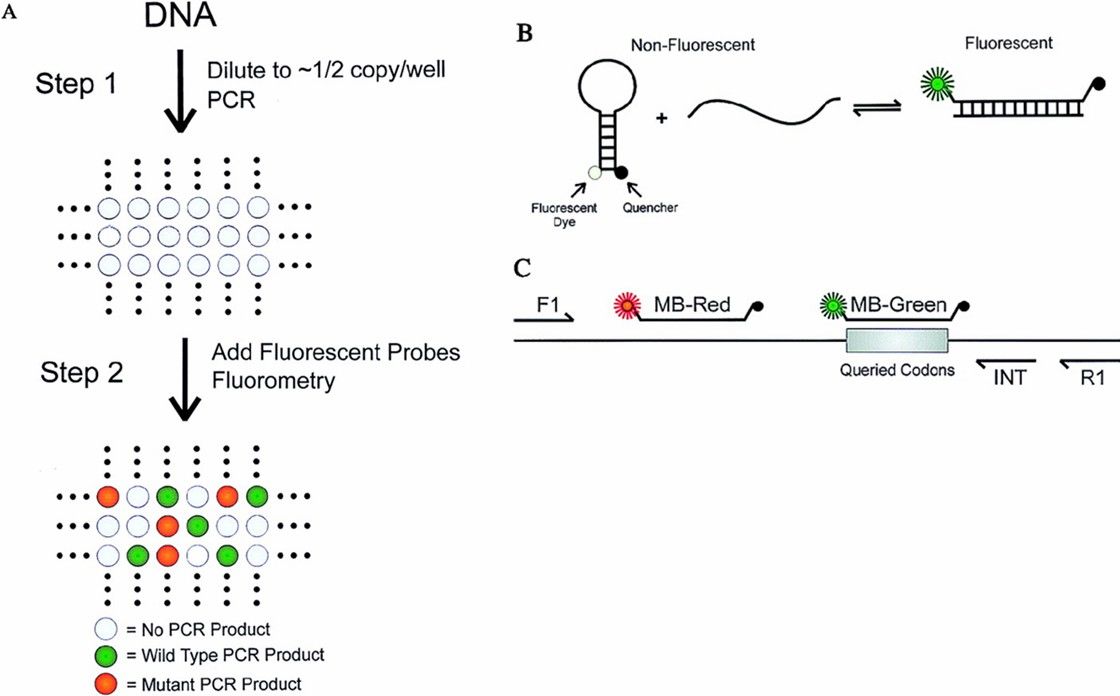 Schematic of Dig-PCR. (Vogelstein, Bert, and Kenneth W. Kinzler. 1999)
Schematic of Dig-PCR. (Vogelstein, Bert, and Kenneth W. Kinzler. 1999)
(4) Direct PCR Method:
In 2023, a new qPCR method called EpiDirect® was introduced, allowing direct methylation quantification in untreated DNA using specially designed INA® (Intercalating Nucleic Acid) primers. EpiDirect® can quantify methylation without bisulfite conversion, with higher sensitivity and specificity. It's demonstrated for analyzing MGMT promoter methylation in glioblastoma patients, though it requires careful primer design and high annealing temperatures (e.g., 76°C).
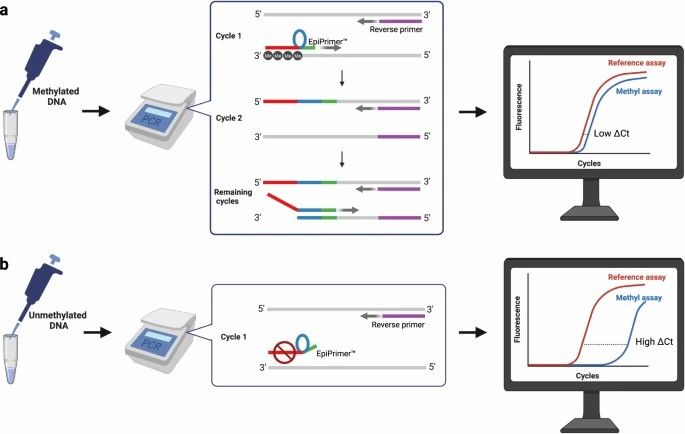 The methyl-specific assay (methyl assay) utilizes an EpiPrimer™ that consists of three parts: An anchor sequence (red), a loop sequence (blue), and a starter sequence (green). (Bendixen, K.K., et al., 2023)
The methyl-specific assay (methyl assay) utilizes an EpiPrimer™ that consists of three parts: An anchor sequence (red), a loop sequence (blue), and a starter sequence (green). (Bendixen, K.K., et al., 2023)
(5) Restriction Enzyme Digestion:
This method uses methylation-sensitive restriction enzymes to digest methylated regions, followed by DNA fragment analysis. A classic enzyme pair is HpaII-MspI, recognizing the CCGG sequence. By using Southern blot or PCR to amplify target fragments, methylation status is determined.
(6) High Resolution Melting (HRM):
Initially for SNP genotyping, HRM is used to detect methylation changes in bisulfite-treated DNA by analyzing differential melting curves with specific dyes. Differences between methylated and unmethylated DNA allow estimation of methylation status by comparing with standard curves.
(7) Affinity Enrichment:
Affinity-based methylation detection includes MeDIP (Methylated DNA Immunoprecipitation) and MBD Cap (Methyl-CpG Binding Domain Capture). MeDIP enriches methylated fragments using antibodies, while MBD Cap employs binding proteins. MeDIP targets low-density, and MBD Cap targets high-density CpG methylation regions.
(8) MassArray:
The MassARRAY EpiTYPER system combines MassCLEAVE (base-specific cleavage) with MALDI-TOF mass spectrometry principles. Following bisulfite treatment, unmethylated cytosine converts to uracil, differentiating molecular weight from methylated cytosine. This system offers high throughput, low cost, and high sensitivity, but requires specific equipment and sample throughput considerations.
Service you may intersted in
Research Strategies in DNA Methylation Studies
The study of DNA methylation modifications typically involves systematic approaches that integrate various molecular biology techniques to elucidate epigenetic regulation mechanisms:
Conventional Research Approach:
Case: Effects of TET2 on DNA Methylation and Leukemia Cell Growth
- Publication: Nature Cell Biology
- Impact Factor: 17.3,
- Publication Date: September 2024.
- Omic Techniques Applied: WGBS for methylation, ChIP-seq for protein-DNA interactions, transcriptomics for gene expression, pulldown proteomics for protein complexes.
- Main Findings: This study highlights the critical role of TET2 in regulating precise DNA demethylation and gene transcription to support tumor growth. In MOLM-13 leukemia cells expressing TET2CDΔLCI, a decline in genome-wide DNA methylation was observed, consistent with non-specific DNA demethylation due to loss of TET2 condensation function in mESCs. This hypomethylation led to altered gene expression, especially upregulation in genes related to cell death and stress response, confirmed by transcriptomic analysis. Correlating with TCGA AML datasets, 15 prognostic biomarkers predicting AML survival were identified. These results suggest that loss of TET2 function alters the 3D genomic structure and transcriptional landscape, impacting clinical outcomes for AML patients.
Integration of Multiplatform Data (DNA Methylation + Transcriptomics)
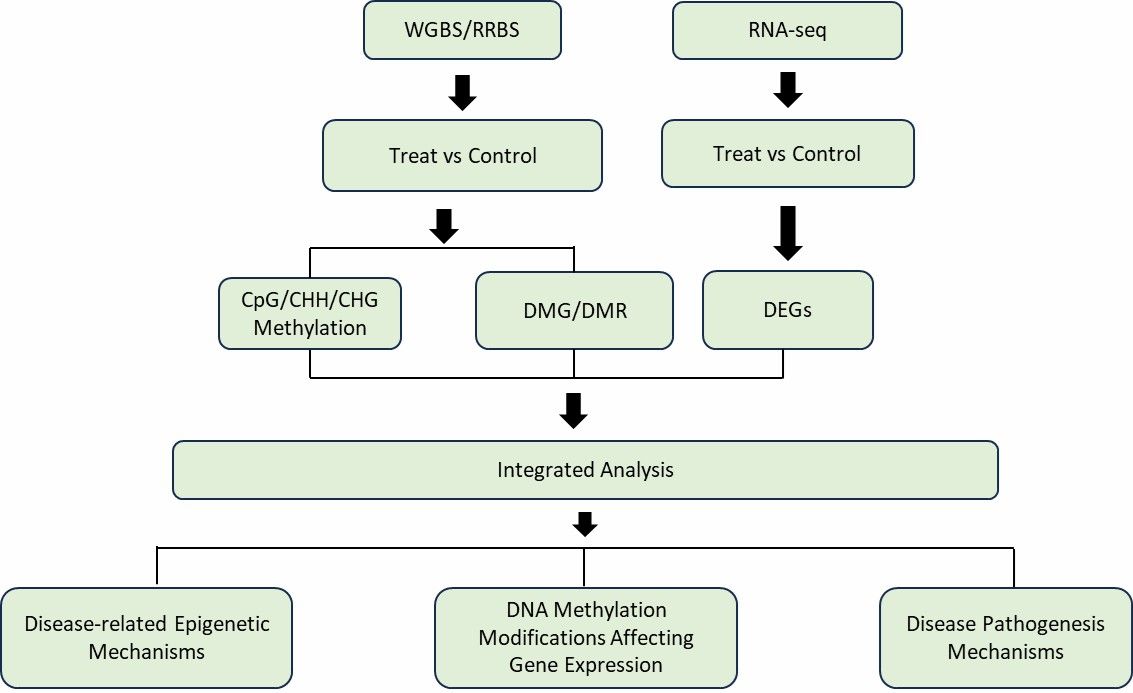
DNA Methylation Modifications Integrated with Multi-Omics The integration of transcriptomic and methylation sequencing data can be divided into:
1. Correlation between transcriptomic data and methylation sequencing data based on genes;
2. Using genes as a bridge to integrate the two omics datasets.
There is a complex regulatory relationship between DNA methylation and gene expression. By correlating gene expression with DNA methylation, we can further understand whether there is an overall correlation between DNA methylation modifications and RNA expression in the samples. Additionally, it helps identify which genes' transcriptional expression is influenced by DNA methylation modifications, and how methylation-regulated genes affect downstream functions.
Case: Foetal hypoxia impacts methylome and transcriptome in developmental programming of heart disease
- Journal: Cardiovascular Research
- Impact Factor: 13.081
- Publication Date: September 2019
- Omics Involved: WGBS, Transcriptome Sequencing
- Abstract: Methylation analysis highlights a significant decline in CpG methylation centered around transcription start sites (TSS). Compared to CGI and CGS located within gene bodies, the CpG islands (CGI) and CpG shores (CGS) within 10 kb upstream of TSS are under-methylated. Integrative analysis with transcriptome data reveals a negative correlation between gene expression around TSS and CpG methylation. Notably, prenatal hypoxia induces opposing methylation patterns in fetal and adult hearts, with hypermethylation observed in fetal hearts and hypomethylation in adult hearts. Furthermore, changes in gene expression patterns in adult hearts exhibit considerable sex-specific differences. Pathway analysis indicates a pronounced enrichment of inflammation-related pathways in the adult male heart compared to the female heart.
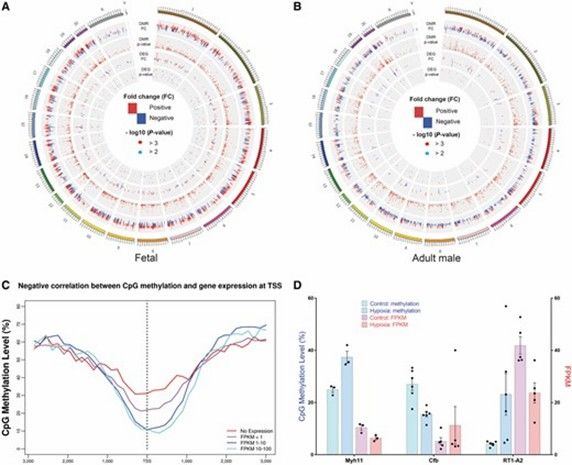 Integration of DMRs and DEGs. (Huang, Lei, et al., 2019)
Integration of DMRs and DEGs. (Huang, Lei, et al., 2019)
Conclusion
In the realm of DNA methylation studies, dynamic regulation of gene expression plays a pivotal role in embryonic development, oncogenesis, and disease mechanisms. Various detection methodologies are employed, each with distinct advantages:
- Whole Genome Sequencing (WGS): This includes methods like WGBS and nanopore sequencing, providing single-base precision for comprehensive analysis.
- Targeted Analysis: Techniques such as pyrosequencing and digital PCR are used, with digital PCR notably supporting the detection of ultra-low abundance targets.
- Methylation Microarrays: Platforms such as Illumina's 850K/935K arrays are ideal for large-scale cohort studies.
- Mass Spectrometry (LC-MS/MS): Offers robust quantification of methylation.
Overview of DNA Methylation Detection Methods
| Detection Type | Method | Key Features | Advantages | Disadvantages | Applications |
| Global Methylation | LC-MS/MS | Digests DNA into nucleotides; quantifies 5mC/5hmC via mass spectrometry. | Gold standard for accuracy/sensitivity. | High cost, complex workflow. | Whole-genome 5mC/5hmC quantification. |
| ELISA | Antibody-based 5mC detection with standard curve. | Fast, low cost, commercial kits available. | Limited sensitivity for subtle changes. | Preliminary global methylation screening. | |
| Sequencing-Based | WGBS | Bisulfite conversion + NGS; single-base resolution. | Full genome coverage, avoids SNP/repeat bias. | High cost, DNA degradation risk. | Pan-cancer methylation profiling (e.g., pancreatic cancer subtypes). |
| oxBS-seq | Chemical oxidation (KRuO₄) + bisulfite treatment to distinguish 5mC/5hmC. | Single-base resolution for 5hmC. | Requires paired BS-seq data. | Hydroxymethylation studies. | |
| RRBS | Restriction enzyme (MspI) + bisulfite sequencing of CpG-rich regions. | Cost-effective for high-CpG regions. | Limited to CpG islands/promoters. | Large clinical cohorts (e.g., cancer biomarker discovery). | |
| scWGBS | Linear amplification + single-cell library prep. | Single-cell resolution, low input. | Technically challenging, low throughput. | Rare cell analysis (e.g., tumor heterogeneity). | |
| Nanopore Sequencing | Direct methylation detection via electrical signal changes. | Long reads, no bisulfite conversion. | High cost, data analysis complexity. | Structural variant-associated methylation. | |
| Microarrays | Infinium (450K/850K/935K) | Bisulfite-converted DNA hybridized to CpG probes; β-value calculation. | High throughput, FFPE-compatible. | Human-specific, limited resolution. | Population epigenome-wide association studies (EWAS). |
| Gene-Specific | MSP/qMSP | Methylation-specific primers for PCR amplification. | Simple, rapid qualitative/semi-quantitative analysis. | Primer design complexity, false positives. | Clinical validation (e.g., Septin9 in colorectal cancer). |
| MethyLight | Probe-based qPCR with FAM/VIC fluorescence. | Quantitative, high specificity. | Requires bisulfite conversion. | Targeted methylation validation (e.g., MGMT in glioblastoma). | |
| Digital PCR (dPCR) | Absolute quantification via microfluidic partitioning. | Ultra-sensitive (0.03%), inhibitor-resistant. | Limited multiplexing capacity. | Liquid biopsy (e.g., cfDNA methylation in early cancer detection). | |
| EpiDirect® | INA® primers for direct methylation detection without bisulfite. | No DNA conversion, high specificity. | Requires high annealing temperatures. | Direct methylation analysis in untreated DNA. | |
| Specialized Assays | MeDIP/MBD-Cap | Antibody/protein-based enrichment of methylated DNA. | Low-input compatibility, broad coverage. | Resolution limited to enriched regions. | Genome-wide methylation patterns (e.g., hypo/hypermethylation in cancer). |
| MassARRAY | Bisulfite PCR + MALDI-TOF mass spectrometry. | High-throughput, cost-effective for mid-scale studies. | Requires bisulfite conversion. | Multi-gene methylation panels (e.g., 40 CpG sites per reaction). |
CD Genomics offers extensive DNA methylation solutions encompassing whole genome sequencing, methylation microarrays, targeted validation using techniques like MSP and MassARRAY, and single-cell analysis. These services cater to the development of epigenetic drugs and integrative multi-omics research, thereby advancing scientific inquiry in these fields.
References
- Greenberg, M.V.C., Bourc'his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20, 590–607 (2019). https://doi.org/10.1038/s41580-019-0159-6
- Harrison, Alan, and Anne Parle-McDermott. "DNA methylation: a timeline of methods and applications." Frontiers in genetics 2 (2011): 74. https://doi.org/10.3389/fgene.2011.00074
- Zhang, H., Lang, Z. & Zhu, JK. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19, 489–506 (2018). https://doi.org/10.1038/s41580-018-0016-z
- Wang, Sarah S., et al. "Whole-genome bisulfite sequencing identifies stage-and subtype-specific DNA methylation signatures in pancreatic cancer." IScience 27.4 (2024). https://doi.org/10.1016/j.isci.2024.109414
- Becker, Daniel, et al. "BiQ Analyzer HiMod: an interactive software tool for high-throughput locus-specific analysis of 5-methylcytosine and its oxidized derivatives." Nucleic acids research 42.W1 (2014): W501-W507. https://doi.org/10.1093/nar/gku457
- Tan, Li, et al. "Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method." Nucleic acids research 41.7 (2013): e84-e84. https://doi.org/10.1093/nar/gkt091
- Javaran, Vahid Jalali, et al. "Grapevine virology in the third-generation sequencing era: from virus detection to viral epitranscriptomics." Plants 10.11 (2021): 2355. https://doi.org/10.3390/plants10112355
- Li, Y., Melnikov, A.A., Levenson, V. et al. A seven-gene CpG-island methylation panel predicts breast cancer progression. BMC Cancer 15, 417 (2015). https://doi.org/10.1186/s12885-015-1412-9
- Gryzinska, Magdalena, et al. "Comparison of programs to design primers for Methylation-Specific PCR." Romanian Biotechnological Letters 24.3 (2019): 479-484. DOI:10.25083/rbl/24.3/479.484
- Eads, Cindy A., et al. "MethyLight: a high-throughput assay to measure DNA methylation." Nucleic acids research 28.8 (2000): e32-00. https://doi.org/10.1093/nar/28.8.e32
- Vogelstein, Bert, and Kenneth W. Kinzler. "Digital pcr." Proceedings of the National Academy of Sciences 96.16 (1999): 9236-9241. https://doi.org/10.1073/pnas.96.16.9236
- Bendixen, K.K., Mindegaard, M., Epistolio, S. et al. A qPCR technology for direct quantification of methylation in untreated DNA. Nat Commun 14, 5153 (2023). https://doi.org/10.1038/s41467-023-40873-y
- Baheti, S., Kanwar, R., Goelzenleuchter, M. et al. Targeted alignment and end repair elimination increase alignment and methylation measure accuracy for reduced representation bisulfite sequencing data. BMC Genomics 17, 149 (2016). https://doi.org/10.1186/s12864-016-2494-8
- Vaisvila, Romualdas, et al. "Enzymatic methyl sequencing detects DNA methylation at single-base resolution from picograms of DNA." Genome research 31.7 (2021): 1280-1289. doi: 10.1101/gr.266551.120
- Guo, L., Hong, T., Lee, YT. et al. Perturbing TET2 condensation promotes aberrant genome-wide DNA methylation and curtails leukaemia cell growth. Nat Cell Biol 26, 2154–2167 (2024). https://doi.org/10.1038/s41556-024-01496-7
- Huang, Lei, et al. "Foetal hypoxia impacts methylome and transcriptome in developmental programming of heart disease." Cardiovascular research 115.8 (2019): 1306-1319. https://doi.org/10.1093/cvr/cvy277





