Epigenetic Breakthroughs in Cancer: From Early Diagnosis to Precision Therapy
Cancer treatment is changing from a focus on genes to a focus on how genes are turned on and off. Normal cells and cancer cells are different. This makes it hard for traditional chemotherapy and targeted therapies to work. Epigenetics is a new breakthrough that can help. It can control changes to DNA and proteins in cells. These changes can be reversed. Gene mutations are permanent, but epigenetic regulation can be reversed, which makes it possible to change tumor cells.
In recent years, major advances in the study of epigenetics at the level of individual cells and in the development of technologies for delivering drugs to specific parts of the body have led to significant progress in the use of drugs for treating a wider range of cancers. For example, a special type of delivery system that carries drugs into cells can make the drugs more concentrated in lung cancer tissues. In addition, drugs can be released in a targeted way using light. For example, liposomal drug delivery systems can precisely increase the concentration of drugs in lung cancer tissues, and light-controlled epitopes can be released locally to minimize side effects. In addition, new drugs (like Aerostat, which targets EZH2) and CRISPR-dCas9 editing tools can target specific areas, which helps them work better than traditional drugs. These tools can help fight chemoresistance and boost the immune system.
In this paper, we systematically analyze the core strategies and clinical challenges of cancer epigenetic therapy.
Pharmacological Targeting Strategies
Cancer epigenetics overcome conventional treatment resistance by dynamically regulating reversible processes such as DNA methylation and histone modification to reshape the tumor gene expression network. In contrast to the irreversibility of gene mutation, the plasticity of epigenetic regulation offers a new opportunity for precise intervention. The significant advancements in single-cell epigenomics and nanomaterial delivery technology have prompted the field to evolve beyond hematological tumors to solid tumors, leading to the realization of "spatio-temporal specific" therapy.
Service you may intersted in
Learn More
Epistatic drug mechanisms: reprogramming engines of gene expression
The primary mechanism of epigenetic drugs involves targeting and regulating epi-modifying enzymes to reverse the abnormal epistatic state of tumor cells. The drugs currently in clinical use are divided into two main categories:
- DNA methyltransferase inhibitors (DNMTi): such as certain nucleoside analogs, by inhibiting DNMT activity, demethylate silencing of the promoter region of oncogenes (e.g., p16, MLH1) and restore their transcriptional function. Clinical studies have shown that low-dose DNMTi treatments improve the 5-year survival rate of patients with myelodysplastic syndromes to 60%.
- Histone deacetylase inhibitors (HDACi): such as first-generation HDAC-targeting agents, activate the expression of differentiation-associated genes (e.g., p21) by blocking HDAC-mediated chromatin compression and increasing histone acetylation levels. These drugs also deregulate tumor stem cells' pluripotency and enhance chemosensitivity.
In contrast to conventional chemotherapy, epimedicines do not directly kill cells. Rather, they induce tumor cell differentiation or immunogenic death through "epigenetic reprogramming." For instance, DNMTi activates endogenous retroviral components and triggers the interferon signaling pathway, while HDACi upregulates tumor antigen presentation and enhances the efficacy of PD-1/PD-L1 inhibitors. The new generation of drugs is moving towards high selectivities, such as the variant EZH2-targeting inhibitors, which can precisely block H3K27me3 modification for the treatment of follicular lymphoma, and CRISPR-dCas9-based epitope editing tools, which can target specific gene sites for methylation or acetylation modification to avoid global epitope perturbation.
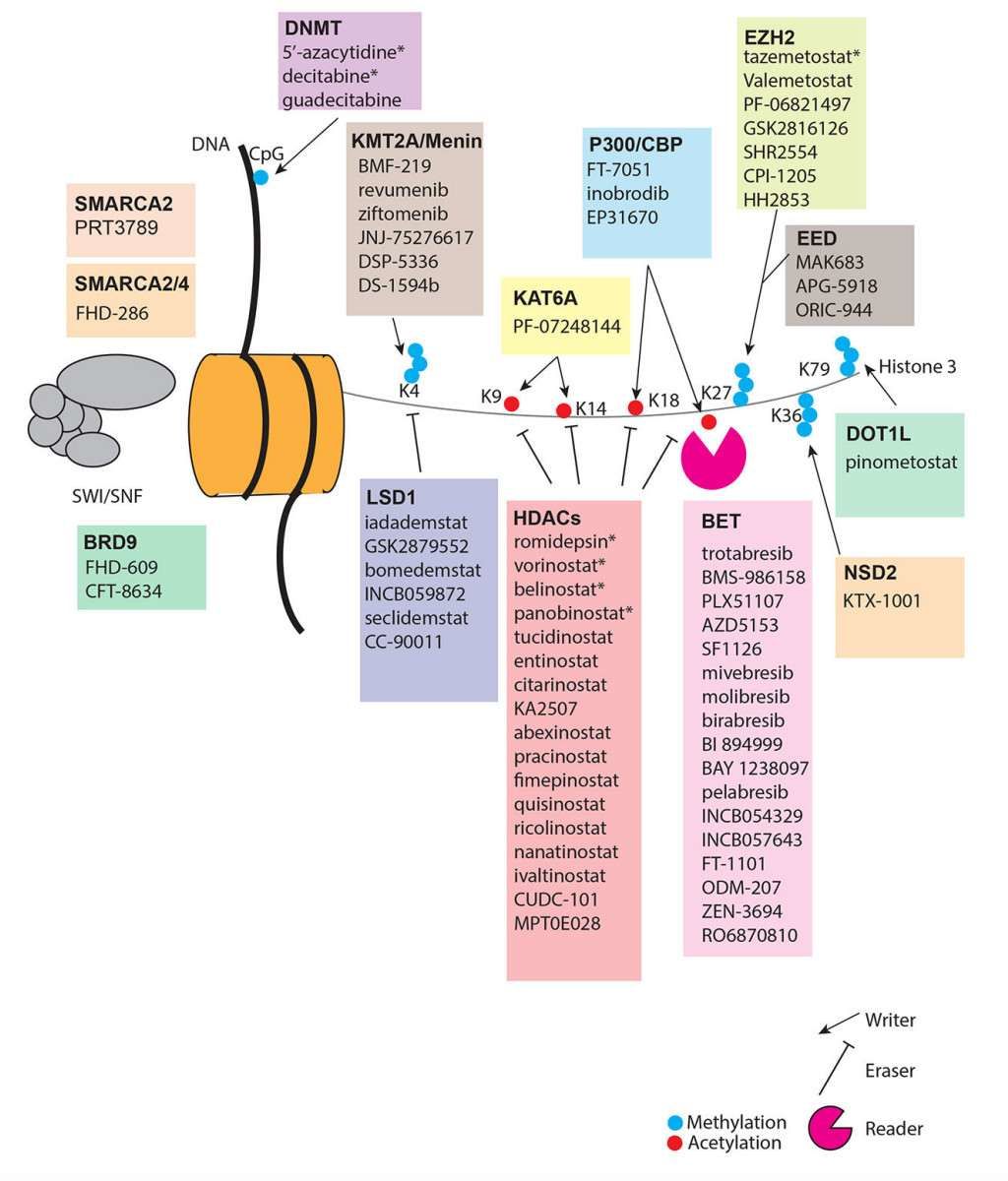 Epigenetic targets in cancer (Mabe et al., 2024)
Epigenetic targets in cancer (Mabe et al., 2024)
Solid tumor therapy: breaking the microenvironmental and Heterogeneity barriers
Despite the effectiveness of epigenetic therapies in leukemia, the complex biology of solid tumors poses multiple challenges in the following three areas:
- Physical barrier: dense stroma with abnormal vasculature impedes drug penetration, e.g. collagen deposition in pancreatic cancer reduces the intratumor concentration of DNMTi.
- Metabolic barrier: hypoxic microenvironment upregulates HDAC6 via HIF-1α, which promotes tumor stem cell epigenetic remodeling and chemoresistance.
- Heterogeneity barrier: Single-cell sequencing has shown that subclones within the same tumor may exhibit distinct methylation patterns, leading to differences in treatment response.
To address these challenges, current research focuses on three main strategies:
- Intelligent delivery systems: liposome-encapsulated epigenetic inhibitors can target lung cancer tissues with a 5-fold increase in intratumor concentration compared to the free drug and a significant reduction in cardiotoxicity.
- Epi-immune combination therapy: HDAC-targeting compounds combined with immune checkpoint inhibitors enhance T-cell infiltration and antigen presentation for synergistic anti-tumor effects in melanoma.
- Spatio-temporal specific intervention: photo-controlled epigenetic compounds can be released locally under near-infrared irradiation, reducing off-target damage to normal tissues.
Preclinical studies have shown that a class of BET protein degraders, an epigenetic degrader, achieved tumor regression in a triple-negative breast cancer model, and liquid biopsy techniques based on circulating tumor DNA (ctDNA) methylation profiles have been used to screen colorectal cancer patients who may benefit from epigenetic therapy.
Epigenomic Biomarkers in Diagnostics
Early diagnosis of cancer is a core component for improving prognosis, and epigenetic markers, especially DNA methylation, have become a key tool for overcoming traditional diagnostic challenges due to their detectability and stability in the early stages of tumorigenesis. In comparison with genetic mutations, methylation modifications are dynamically reversible and closely associated with the tumor microenvironment, providing unique advantages for non-invasive screening. For instance, fecal and blood-based methylation detection technologies (e.g., multi-target stool and plasma assays) have been clinically validated for colorectal cancer, marking the transition of epigenetics from basic research to precision medicine.
Service you may intersted in
Clinical Breakthroughs in Methylation Detection Technology
Abnormal DNA methylation is an early event in cancer development, which drives tumor progression by silencing oncogenes (e.g., p16, MLH1) or activating oncogenic pathways. Methylation detection techniques currently used in clinical applications fall into two main categories:
- Cologuard (Stool Test): Cologuard enables multi-dimensional screening for colorectal cancer by jointly analyzing the methylation of the BMP3 and NDRG4 genes, KRAS mutations, and hemoglobin in feces. It has a sensitivity of 92% and nearly doubles the detection rate of precancerous lesions compared to the traditional fecal occult blood test (FIT). However, the false-positive rate of approximately 13% can lead to overmedication and sample collection is susceptible to dietary and intestinal flora interference.
- Epi proColon (blood test): Epi proColon targets the methylation site of the Septin9 (SEPT9) gene in plasma and enhances assay stability with a single-tube, dual-target design (detecting both the mSEPT9 and ACTB genes). With a sensitivity of 94% and specificity of 84%, it is particularly suitable for people for whom colonoscopy is contraindicated. The new generation Epi proColon 2.0 further optimizes performance by integrating markers such as mBCAT1 to address the technical bottleneck of low abundance of circulating tumor DNA (ctDNA) in blood.
Current technological trends include multi-omics coupling (e.g., methylation + protein markers) and ctDNA enrichment with nanomaterials, which are aimed at further improving detection sensitivity. In addition, pan-cancer assays (e.g., GRAIL's Galleri test) are exploring the feasibility of "screening multiple cancers with one tube of blood" in an attempt to cover the methylation profiles of multiple tumors with a single assay.
Liquid Biopsy for Epigenomic Profiling
Dynamic changes in cancer epigenetic regulation (e.g., DNA methylation, histone modifications) are central mechanisms driving tumor heterogeneity and treatment resistance. Traditional tissue biopsies make it difficult to capture the real-time evolution of epigenetic features due to high invasiveness and unrepeatable sampling. Liquid biopsy provides a breakthrough for non-invasive, dynamic monitoring of epigenetic abnormalities by analyzing biomarkers such as circulating tumor DNA (ctDNA). Technologies represented by cfMeDIP-Seq and cfChIP-Seq are reshaping the clinical practice of early cancer diagnosis, monitoring of microscopic residual disease, and precision treatment strategies by resolving methylation patterns and histone modifications.
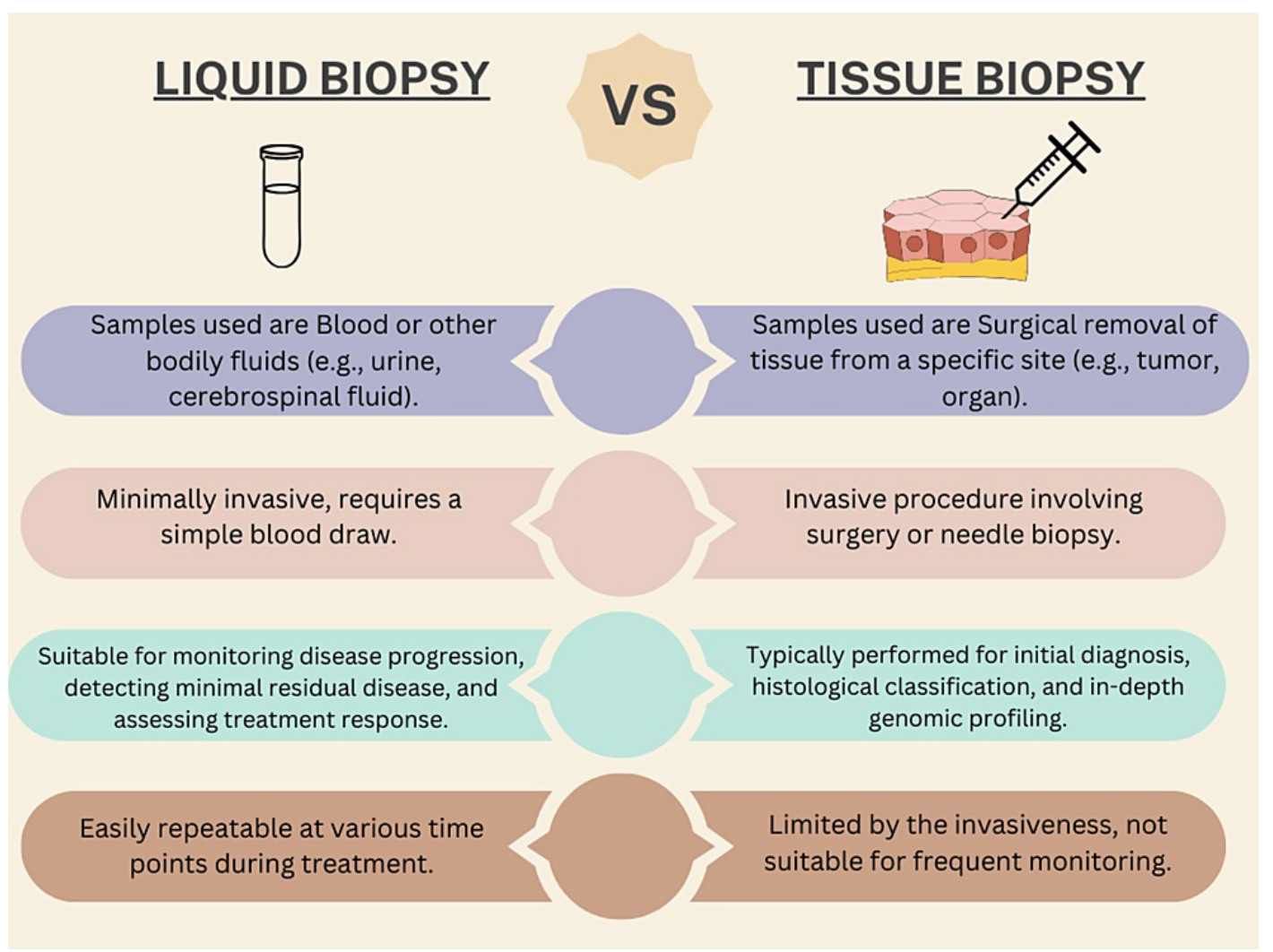 Comparison between liquid biopsy and traditional tissue biopsy (Adhit et al., 2023)
Comparison between liquid biopsy and traditional tissue biopsy (Adhit et al., 2023)
Non-invasive epigenetic analysis techniques
The core advantage of liquid biopsy is its non-invasive and dynamic monitoring capability. By capturing free DNA (cfDNA) in the blood, cfMeDIP-Seq and cfChIP-Seq technologies are able to resolve epigenetic features of tumors and provide highly sensitive molecular information for clinical use.
- cfMeDIP-Seq: This technology utilizes methylation-specific antibodies to enrich for methylated DNA fragments, which, combined with high-throughput sequencing, enables genome-wide methylation mapping. For example, in early-stage lung and colorectal cancers, cfMeDIP-Seq has enabled the detection of ctDNA abundance as low as 0.1% with a sensitivity of over 90%. Its clinical value is particularly prominent in pan-cancer screening (e.g., GRAIL's Galleri test), where combinations of methylation signatures can be traced back to tumor tissue origin.
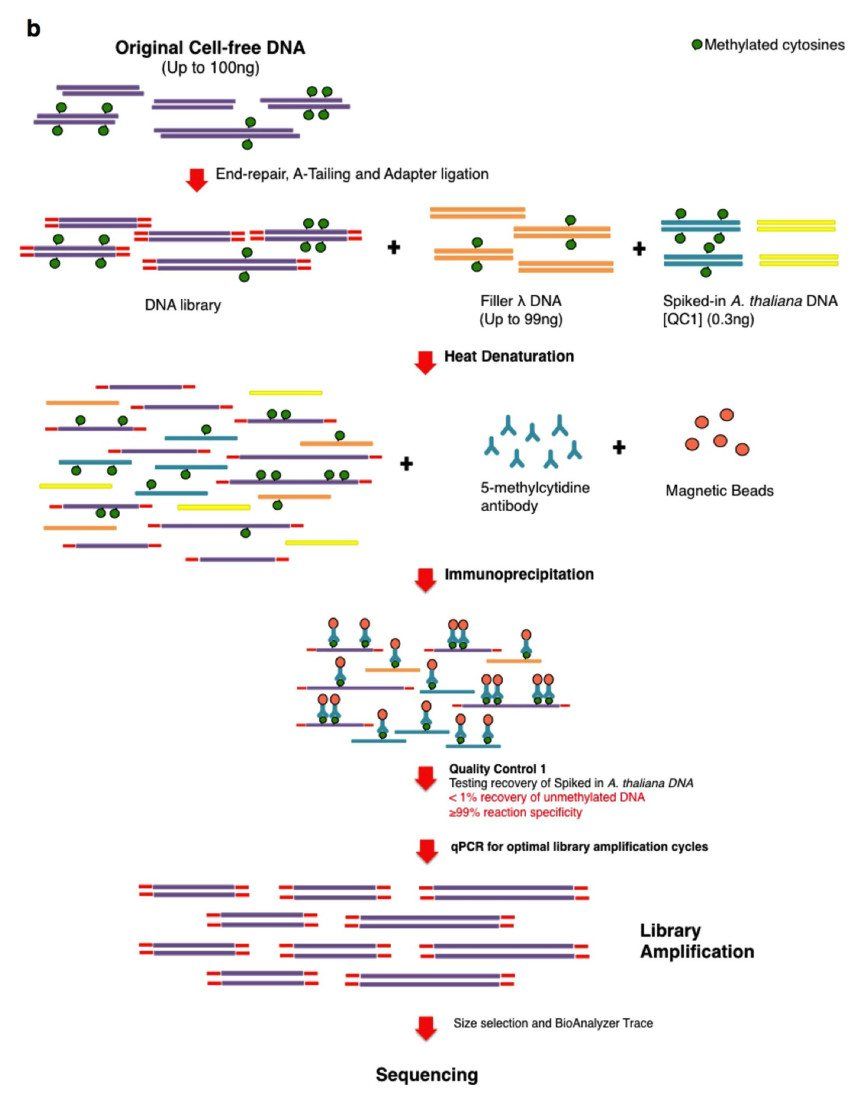 Schematic representation of the cfMeDIP–seq protocol (Shen et al., 2018)
Schematic representation of the cfMeDIP–seq protocol (Shen et al., 2018)
- cfChIP-Seq: By targeting histone modifications (e.g. H3K27ac, H3K4me3) or transcription factor binding sites, cfChIP-Seq can reveal aberrant activation of gene regulatory networks. Recent studies have shown that this technology can detect changes in ERα activity in the blood of breast cancer patients, providing a basis for the prediction of endocrine therapy resistance.
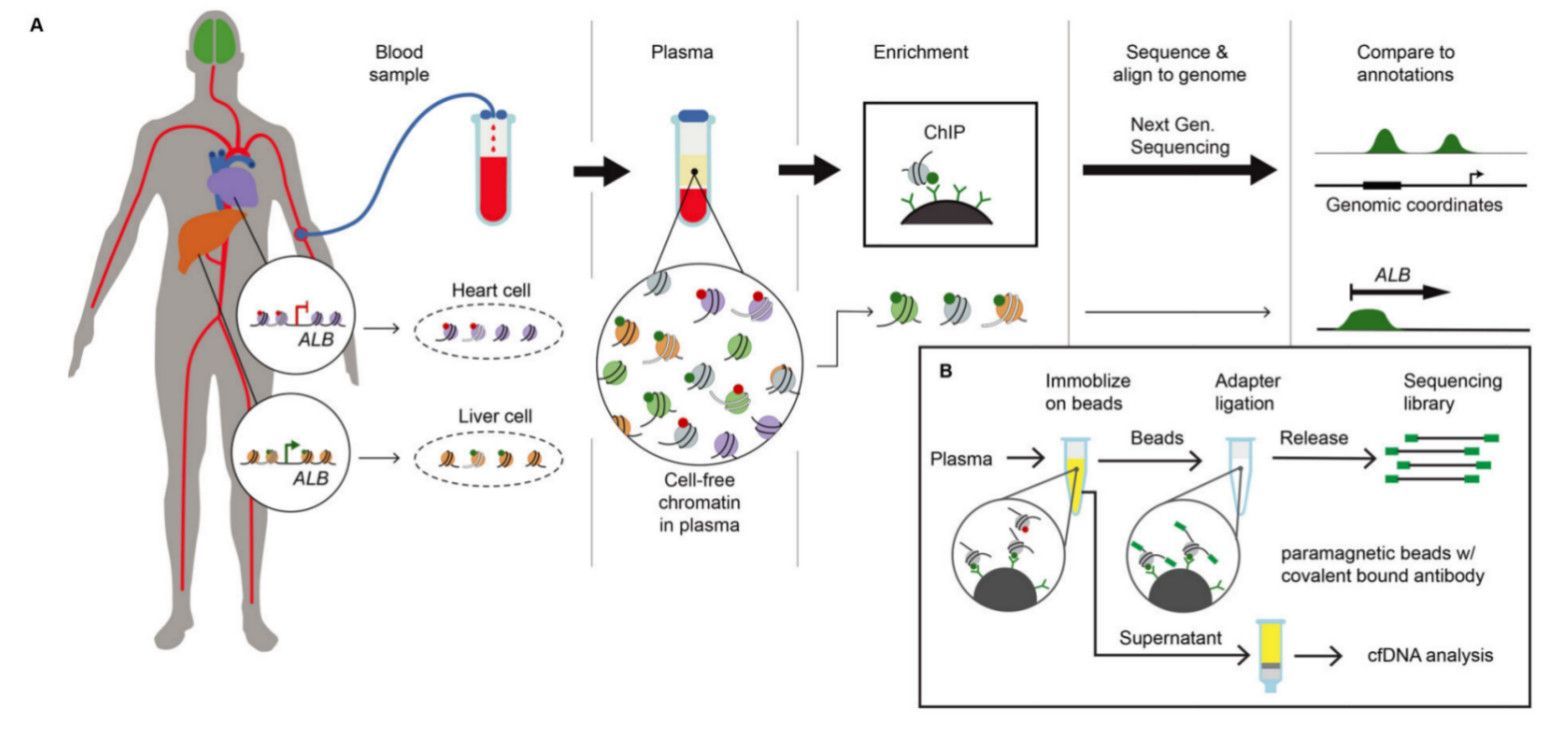 cfChIP-seq method outline and protocol (Sadeh et al., 2021)
cfChIP-seq method outline and protocol (Sadeh et al., 2021)
Current technological developments focus on multi-omics integration (e.g. methylation combined with fragmentomics) and single-cell epigenetic analysis to enhance detection resolution. The application of nanomaterial enrichment and microfluidic chip is dedicated to overcoming the technical bottleneck of low abundance of ctDNA, and to promoting the translation of liquid biopsy from scientific research to clinic.
From microscopic residual disease to the clinical application of precision therapy
The value of liquid biopsies in cancer diagnosis and treatment extends throughout the course of the disease. In microscopic residual disease (MRD) surveillance, ctDNA methylation markers (e.g., SEPT9) can provide an early warning of recurrence risk when imaging is negative. For example, ctDNA-positive patients after colorectal cancer surgery have a 17-fold higher risk of recurrence than negative patients, and cfMeDIP-Seq has a detection limit as low as 0.01% for MRD.
Epigenetic traits (e.g., genome-wide hypomethylation) are strongly associated with tumor aggressiveness. Analysis of H3K4me3 modification levels by cfChIP-Seq predicts progression-free survival in lung cancer patients, while dynamic scoring of ctDNA methylation patterns has been used for lymphoma efficacy assessment. In addition, liquid biopsies can tap therapeutic targets (e.g., DNMT1) and monitor the evolution of drug resistance. HIF2α activity in renal cell carcinoma can be detected by cfChIP-Seq, which can guide the combination of targeted drugs; dynamic changes of EGFR methylation in non-small cell lung cancer correlate with the efficacy of ositinib and provide a basis for treatment adjustment.
Future of Epigenome Editing
CRISPR/dCas9 epigenetic tools are changing cancer treatment. They are moving from passive diagnosis to active intervention. The technology uses a special version of a protein (called "dCas9") that can target specific areas of DNA (like promoters or enhancers) and control which genes are turned on or off. It does this by working with other molecules that can change the DNA's makeup (like DNA methylase or histone acetylase). This process can turn on or off genes that cause cancer (like TP53) or silence genes that cause cancer (like MYC), and all of this happens without cutting the DNA itself. For example, the dCas9-p300 system activates a process called "TP53-induced apoptosis" in lung cancer cells. In another example, dCas9-TET1 reverses the abnormal methylation of CDKN2A in ovarian cancer cells, which restores its oncogenic function. Compared to traditional epigenetic drugs (like demethylating agents), this precision editing strategy overcomes the limitations of older methods and speeds up the process of getting these drugs to patients. It does this by using single-cell epigenomics and integrating multiple types of omics data.
This technology also makes cancer treatment more advanced by changing it from "biomarker-dependent" to "remodeling epigenetic abnormalities." CRISPR/dCas9 can improve the response rate of immunotherapy by targeting genes that cause tumors to escape from the immune system (for example, MHC-I) or by enhancing the function of CAR-T cells. Customized editing strategies for specific patient problems (for example, the ASXL1 mutation in AML) can reverse chemoresistance. Although there are still issues with how well it works, the risks, and the ethics, more than a dozen clinical trials have been conducted around the world to explore its use in blood cancers and solid tumors. In the future, with improvements in delivery systems (like lipid nanocarriers) and dynamic regulation technologies, epigenome editing is expected to become a key tool for precision cancer treatment, making the vision of "editing as therapy" a reality.
References
- Mabe, Nathaniel W et al. "Pharmacological targeting of the cancer epigenome." Nature cancer vol. 5,6 (2024): 844-865. doi:10.1038/s43018-024-00777-2
- Adhit, Kanishk K et al. "Liquid Biopsy: An Evolving Paradigm for Non-invasive Disease Diagnosis and Monitoring in Medicine." Cureus vol. 15,12 e50176. 8 Dec. 2023, doi:10.7759/cureus.50176
- Shen, Shu Yi et al. "Sensitive tumour detection and classification using plasma cell-free DNA methylomes." Nature vol. 563,7732 (2018): 579-583. doi:10.1038/s41586-018-0703-0
- Sadeh, Ronen et al. "ChIP-seq of plasma cell-free nucleosomes identifies gene expression programs of the cells of origin." Nature biotechnology vol. 39,5 (2021): 586-598. doi:10.1038/s41587-020-00775-6




