Cancer Epigenome: From DNA Methylation and Diet to New Frontiers in Cancer Therapy
For decades, cancer was seen primarily as a genetic disease—triggered by DNA mutations that disrupt cell growth. But recent breakthroughs in epigenetics have shifted that view. Scientists now recognise that chemical changes to DNA, such as methylation and histone modification, also play a critical role in how cancer develops, spreads, and responds to treatment.
The key difference? Unlike mutations, epigenetic changes are reversible. That opens new therapeutic doors. For example, DNA methyltransferase (DNMT) inhibitors have shown strong results in blood cancers by reactivating silenced tumour suppressor genes. Similarly, diet-derived compounds like folic acid and polyphenols have been linked to changes in DNA regulation that may reduce cancer risk.
Still, translating epigenetics into real-world therapies isn't without its hurdles:
- Solid tumours present a highly complex microenvironment.
- Many epigenetic drugs, especially stereoisomers, risk unintended effects in healthy cells.
- And every patient is biologically unique—responses vary widely.
To overcome these challenges, research is becoming more holistic:
- Single-cell epigenomics is revealing how individual cells evolve within tumours.
- Nanoparticle drug delivery systems are improving treatment accuracy at the cellular level.
- Artificial intelligence is helping predict how patients might respond to different epigenetic drugs or diet-based interventions.
Together, these innovations are moving cancer research beyond a "gene-first" mindset. Instead, we're entering an era where genetics, epigenetics, and environmental factors are seen as a connected network—one that can be targeted from multiple angles.
This review explores three pillars shaping the future of cancer epigenetics:
- Core regulatory mechanisms
- Scientific evidence supporting nutritional modulation
- Epigenetic-based therapeutic strategies
Cancer Epigenome Overview
Cancer doesn't just develop because of genetic mutations. It's also related to problems in the epigenetic regulatory networks. Epigenetic mechanisms influence cell fate by changing DNA and histones to control gene expression without changing the DNA sequence. Abnormalities in the cancer epigenome are associated with tumor heterogeneity, metastasis, and drug resistance. The fact that these abnormalities can be reversed offers unique opportunities for targeted therapies. In the next section, we will examine how epigenetic inheritance works, especially how it interacts with genetic mutations and its role in regulating oncogenes.
Definition and core features of the cancer epigenome
The "cancer epigenome" refers to the sum of abnormalities in DNA methylation, histone modifications, chromatin remodeling, and non-coding RNA regulation in tumors. Its core features include hypermethylation of CpG islands in gene promoter regions (e.g., p16INK4A silencing), genome-wide hypomethylation (activation of proto-oncogenes such as c-MYC), and chromatin compression mediated by histone modifications (e.g., H3K27me3). These abnormalities remodel the cellular phenotype by altering gene accessibility.
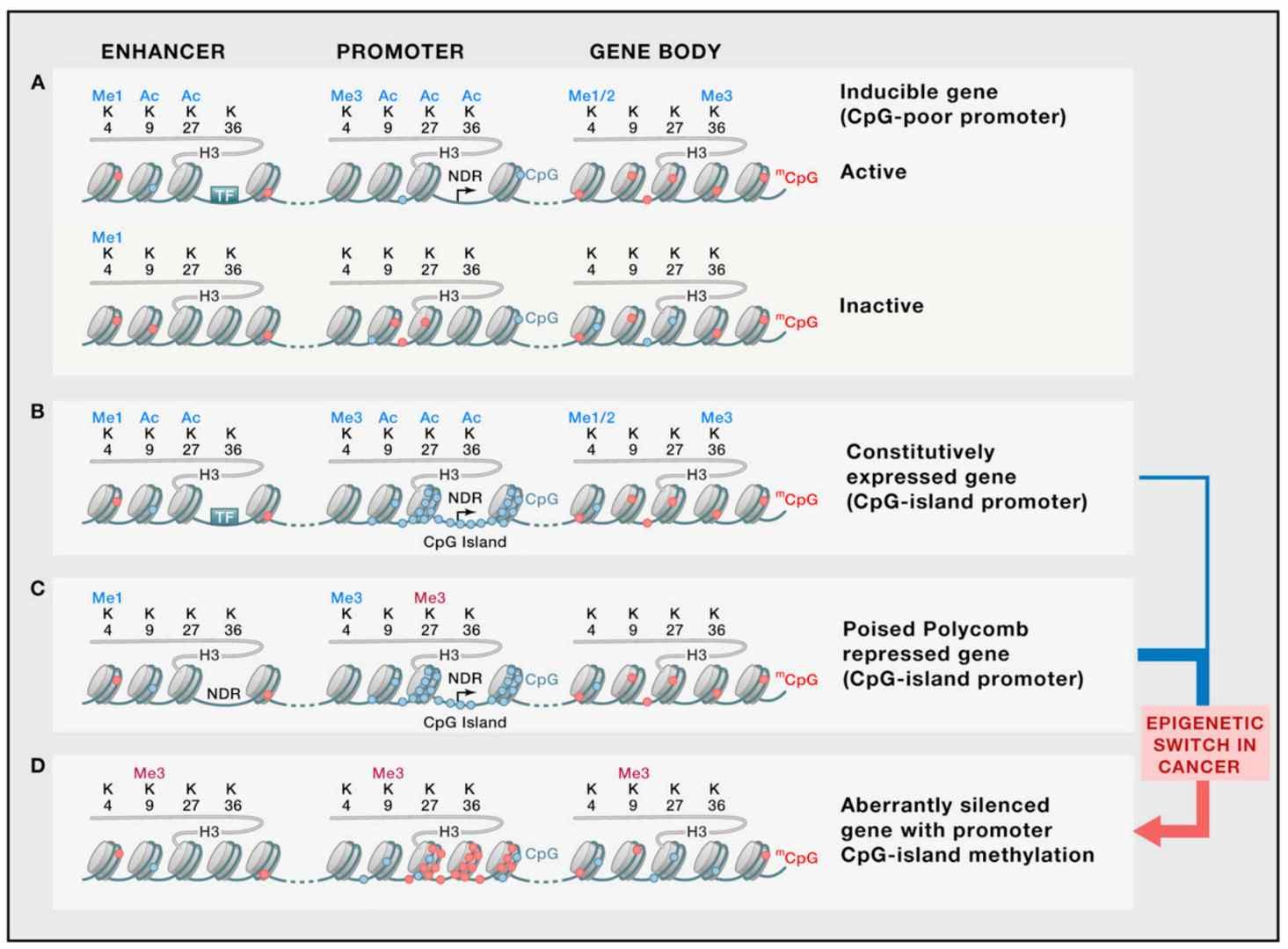 Representative epigenetic states (Shen et al., 2013)
Representative epigenetic states (Shen et al., 2013)
Recent studies have revealed that mutations in epistatic regulators (e.g., DNMT3A, TET2) can directly disrupt methylation patterns, whereas non-coding RNAs (e.g., HOTAIR) promote gene silencing by recruiting chromatin modification complexes. The clinical validation of epigenetic drugs, such as DNA methyltransferase (DNMT) inhibitors, has been demonstrated in the context of hematologic tumors, thereby substantiating the therapeutic potential of epigenetic targets.
Service you may intersted in
Learn More
Synergy and competition between epigenetic and genetic mutations
Genetic mutations, such as TP53 deletion, directly disrupt gene function. In contrast, epigenetic abnormalities indirectly drive tumor evolution by regulating gene expression. The two often act synergistically: for example, MLH1 promoter hypermethylation in colon cancer leads to defective DNA repair and accelerates the accumulation of subsequent mutations; IDH1 mutations inhibit histone demethylase through the metabolite 2-hydroxyglutarate, creating a pro-cancer epigenetic environment.
The reversibility of epigenetic regulation has been identified as a significant therapeutic breakthrough. The administration of low-dose DNMT inhibitors has been demonstrated to induce reprogramming of the episodic landscape, leading to the activation of oncogenes and the enhancement of immune responses. The present research endeavors concentrate on combination therapies (e.g., epimedicines + immune checkpoint inhibitors) and single-cell epigenomic technologies to resolve tumor heterogeneity and optimize precision therapy.
Epistatic mechanisms of oncogene activation and oncogene silencing
Epigenetic aberrations have been shown to play a pivotal role in the process of malignant transformation through a dual pathway. The silencing of oncogenes, such as BRCA1, is predominantly facilitated by promoter hypermethylation or H3K27me3 modification. In contrast, the activation of oncogenes, including MYC, is often associated with enhancer hypermethylation or histone acetylation. Non-coding RNAs (e.g., miR-29 targeting DNMT3A) function as regulatory hubs in this process.
Cutting-edge technologies such as CRISPR-dCas9 epitope editing have been developed for the specific reversal of oncogenic modifications, while the delivery of methylation inhibitors via nanocarriers has led to significant advancements in the targeting of solid tumors. The objective of these strategies is to restore epigenetic homeostasis and impede tumor progression.
Epigenetic Drivers of Cancer
Cancer develops and progresses not only because of genetic mutations but also because of significant disruptions in the epigenetic regulatory network. There are three major mechanisms that change the structure of the genome and the way genes are expressed without altering the DNA sequence: DNA methylation, histone modification, and chromatin remodeling. These mechanisms cause tumors to start, make tumors different from each other, and make tumors resistant to treatment. These epigenetic alterations can either cause cancer on their own or work together with genetic mutations to make cancer more complicated.
Epigenetic core mechanisms
DNA methylation has been observed to exhibit a dual imbalance, characterized by global hypomethylation and local hypermethylation, within neoplastic tissues. Hypomethylation of genome-wide regions (e.g., transposons and repetitive sequences) has been shown to induce chromosomal breaks and activate proto-oncogenes (e.g., c-MYC). In contrast, hypermethylation of promoters of oncogenes (e.g., p16INK4A) has been demonstrated to silence gene expression through the recruitment of methyl-binding proteins (e.g., MeCP2). Recent studies have revealed that mutations in regulatory factors such as DNMT3A and TET2 can reshape tumor methylation profiles, and single-cell techniques have further revealed the impact of methylation heterogeneity on therapeutic resistance.
Histone modifications regulate the status of chromatin through the application of dynamic chemical marks, including acetylation and methylation. For instance, H3K27ac marks active enhancers, thereby promoting oncogene expression. Conversely, PRC2 complex-mediated H3K27me3 results in the compaction of chromatin and the repression of differentiation-related genes. Aberrant expression of histone-modifying enzymes (e.g., EZH2) has been demonstrated to confer stemness to cancer cells, and the clinical success of their inhibitors (e.g., Tazemetostat) validates the therapeutic potential of targeting histone modifications.
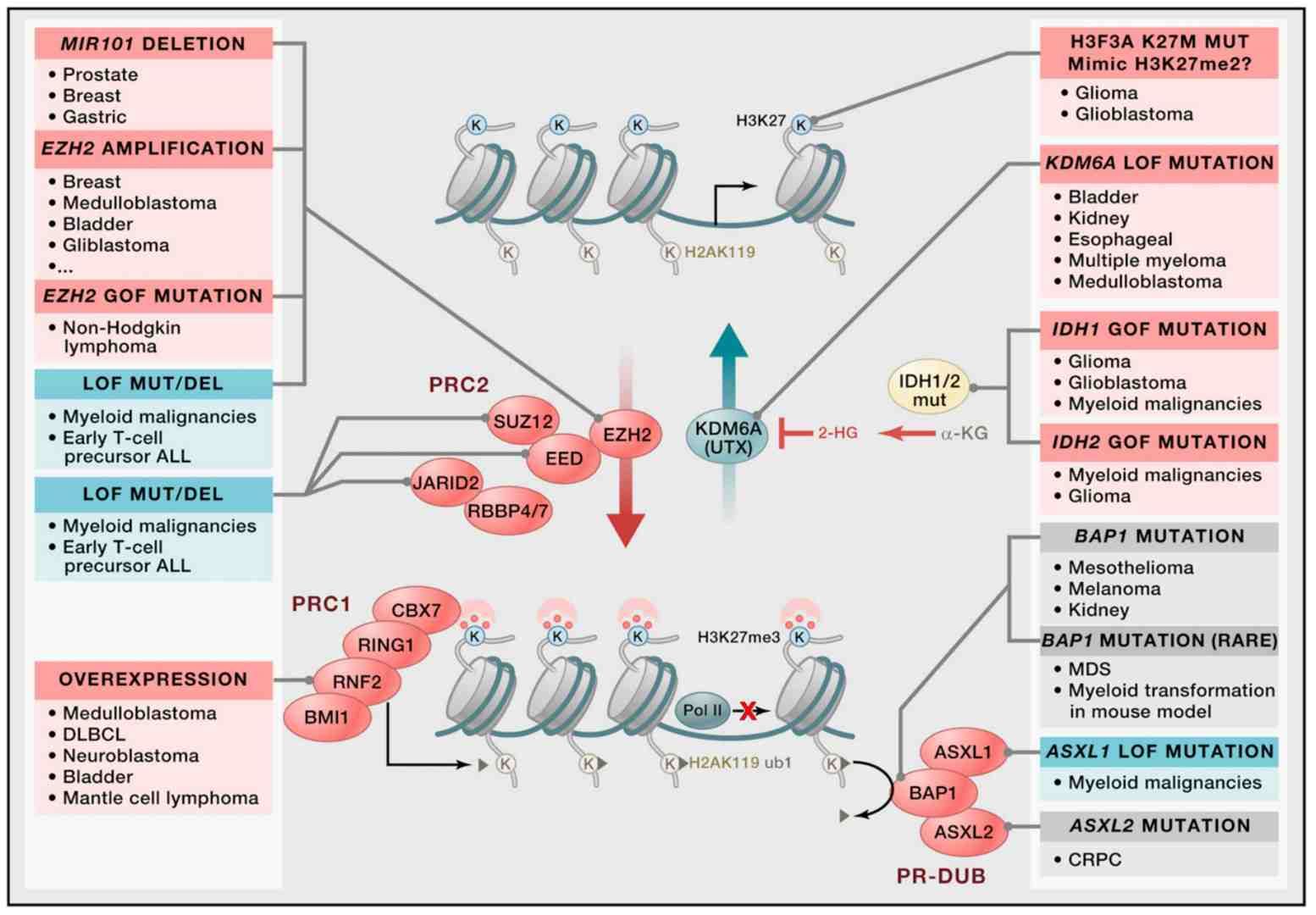 Genetic disruption of epigenetic control at H3K27 in cancer (Shen et al., 2013)
Genetic disruption of epigenetic control at H3K27 in cancer (Shen et al., 2013)
Chromatin remodeling complexes (e.g., SWI/SNF) have been shown to expose or hide critical DNA regions by altering nucleosome positions. Inactivating mutations in genes such as ARID1A have been observed to lead to chromatin loosening and activation of proto-oncogenic enhancers. At the same time, BRG1 deletion has been demonstrated to disrupt DNA repair and exacerbate genomic instability. The advent of CRISPR-dCas9-based epitope editing technology has catalyzed the development of precision intervention strategies.
Epigenetic dynamics and tumor heterogeneity
Epigenetic abnormalities have been shown to precede genetic mutations as a "precursor event" for tumorigenesis. The chronic inflammatory microenvironment has been shown to induce local hypermethylation and silence oncogenes by inhibiting TETase activity. Concurrently, EZH2 overexpression has been observed to promote cellular immortalization by inhibiting the INK4A-ARF pathway. These early alterations create a genomic environment conducive to mutation accumulation.
Tumor heterogeneity is the result of a complex interplay between epigenetic plasticity and the microenvironment. Single-cell epigenomics has been demonstrated to reveal that subpopulations of cells within the same tumor may exhibit distinct methylation or histone modification patterns. For instance, the condition of hypoxia activates histone deacetylases (HDACs) via the hypoxia-inducible factor-1α (HIF-1α), inducing mesenchymal phenotypes and chemoresistance. Conversely, stochastic expression of DNA methyltransferase 3B (DNMT3B) leads to subclonal epigenetic differentiation, creating differences in metastatic potential. This dynamic adaptability enables cancer cells to evade therapeutic pressure, as DNMT inhibitors may selectively enrich resistant cell populations.
The present state of research is oriented towards the investigation of epigenetic drug combination strategies (e.g., DNMT+HDAC inhibitors) and their synergy with immunotherapy. Concurrently, spatial epigenomics techniques are resolving the spatial and temporal impact of the microenvironment on epigenetic regulation. The integration of single-cell multi-omics and artificial intelligence modeling in the future is expected to achieve precise resolution and intervention of heterogeneity.
Service you may intersted in
Learn More
Epigenetic Diet Influence
The connection between diet and cancer isn't just about how energy is processed. It also has a significant impact on how tumors form and grow through epigenetic regulatory networks. Nutrients such as folic acid, choline, polyphenols, and others have been shown to interfere with oncogene activation and silencing by regulating DNA methylation, histone modification, non-coding RNA expression, and reshaping gene expression profiles. Nutritional epigenomics is a new field that studies how what we eat affects cancer risk. It has helped scientists create a plan for how to prevent cancer more effectively. This section will focus on how diet affects gene activity and the studies that support this.
Epigenetic regulatory mechanisms of dietary composition
Folate, a pivotal component of one-carbon metabolism, plays a critical role in maintaining DNA synthesis and methylation homeostasis by supplying methyl groups. Its deficiency has been shown to result in extensive genome hypomethylation and induce chromosomal instability. In addition, hypermethylation of specific oncogenes (e.g., p16INK4a) has been observed to silence their expression. Choline, conversely, has been demonstrated to maintain methylation homeostasis by supporting S-adenosylmethionine (SAM) synthesis. A deficiency of choline during pregnancy has been shown to alter histone modifications associated with fetal neurodevelopment (e.g., H3K9me2) and to correlate with an increased risk of disease in adulthood. Polyphenols (e.g., curcumin, and resveratrol) reverse the silencing of oncogenes by inhibiting DNMT and HDAC activity. However, their low bioavailability has prompted researchers to develop nano-delivery systems to enhance efficacy.
In a global epigenetics study of dietary patterns, a Mediterranean diet high in polyphenols and omega-3 fatty acids was found to reduce colorectal cancer risk by inhibiting DNA methylation of pro-inflammatory genes (e.g., TNF-α) and enhancing histone acetylation. Conversely, a Western diet characterized by high consumption of red meat and low fiber intake has been associated with a 3.25-fold elevated risk of cancer recurrence. This association is hypothesized to occur through a mechanism involving episodic activation of the TLR4 pathway, driven by dysbiosis of the intestinal flora. Conversely, caloric restriction (CR) has been shown to impede tumor epigenetic senescence by activating SIRT1 deacetylase and inhibiting DNMT activity. These findings imply that dietary patterns can influence long-term cancer susceptibility through a process known as epigenetic "memory." Nutrients and dietary patterns act as "molecular switches" in cancer prevention by modulating methyl donor networks and epimerase activity. However, the effects of these nutrients are subject to regulation by genetic polymorphisms and metabolic status.
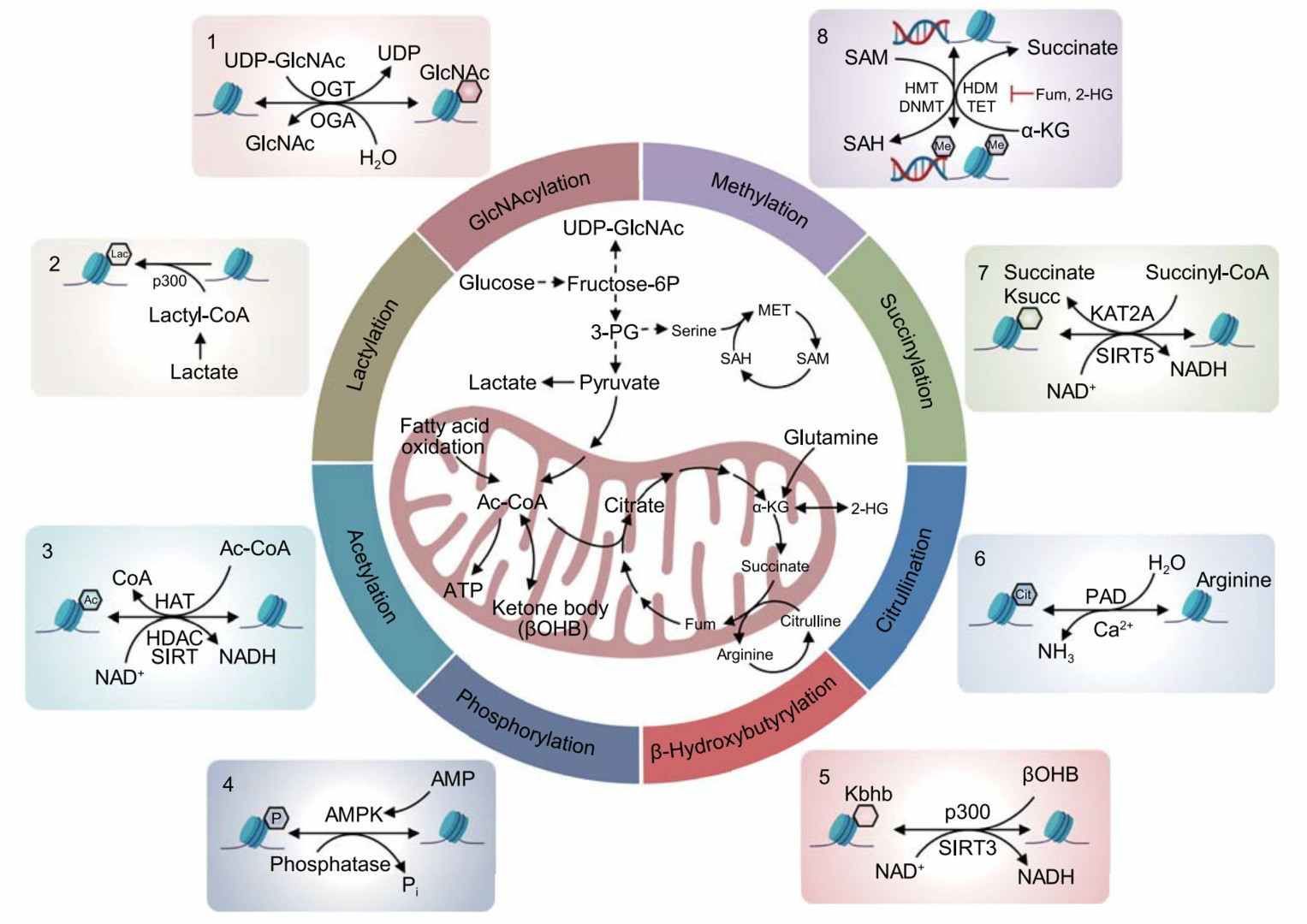 An overview of metabolic connections to epigenetic remodeling (Sun et al., 2022)
An overview of metabolic connections to epigenetic remodeling (Sun et al., 2022)
From lab to population: research evidence and translational challenges
Mechanistic validation in preclinical models demonstrated that folic acid deficiency induced abnormal H3K27me3 modification in the embryonic neural tube in animal experiments, and folic acid supplementation partially reversed the methylation disorder. Furthermore, choline supplementation during pregnancy elevated the DNA methylation level in the hippocampal region of the offspring and improved their cognitive function. In a hepatocellular carcinoma model, curcumin was found to restore p53 activity and reduce tumor volume by inhibiting DNMT3B. These findings provide molecular evidence for dietary intervention; however, species differences and dose effects require further interpretation with caution.
A causal exploration of population-based studies demonstrated a 30% reduction in all-cause mortality among older adults who consumed more than 650 mg of polyphenols per day, primarily from fruit and coffee, in the Italian cohort study. Additionally, a significant increase in the 5-year risk of recurrence was observed in the High Western Dietary Patterns group among patients with stage III colon cancer. Single-cell epigenomic techniques have further revealed that dietary interventions can remodel the methylation heterogeneity of immune cells in the tumor microenvironment and enhance chemosensitivity. However, individual genetic backgrounds (e.g., MTHFR C677T polymorphism) and intestinal flora differences lead to fluctuations in intervention effects, highlighting the need for personalized nutritional strategies. A synthesis of preclinical and population studies substantiates the epimodulatory capacity of diet. Nevertheless, individual heterogeneity and technical limitations persist as significant impediments to clinical implementation.
Epigenetic Cancer Therapies
In animal experiments, a lack of folic acid caused problems with a specific change (H3K27me3 modification) in the developing nervous system. Adding folic acid to the animals' diets partly fixed these problems. In addition, when pregnant women took choline supplements, it increased the levels of DNA methylation in the hippocampus (a part of the brain) of their babies. This led to improved cognitive function (thinking, learning, and memory) in the babies. In a study of liver cancer, curcumin was found to help p53 work better and reduce tumor size by stopping a process called DNA methylation. These findings provide molecular evidence that a person's diet can help with weight loss, but more research is needed because of differences between species and the effects of different doses.
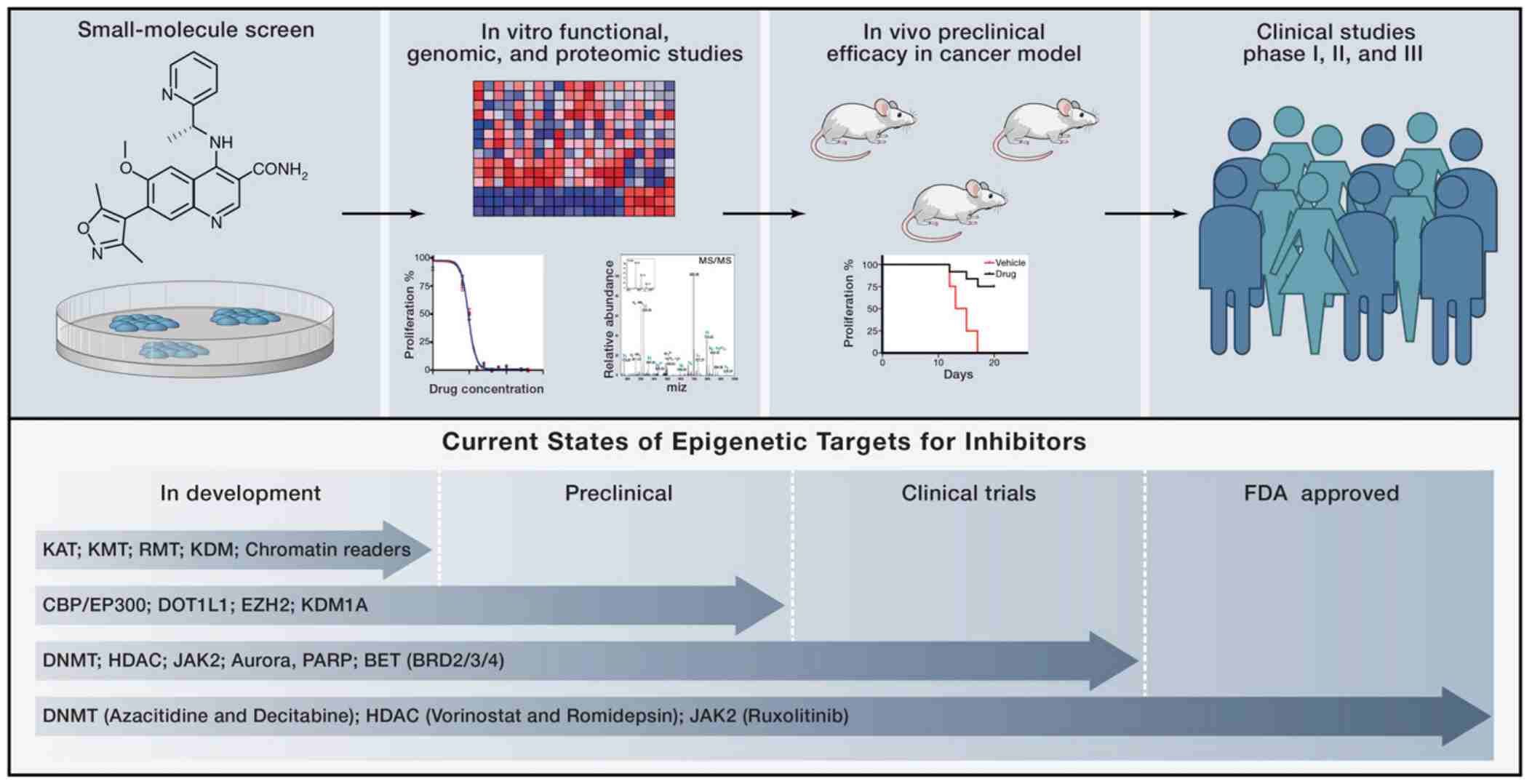 Epigenetic Inhibitors as Cancer Therapies (Dawson & Tony, 2012)
Epigenetic Inhibitors as Cancer Therapies (Dawson & Tony, 2012)
Researchers found that older adults who consumed more than 650 mg of polyphenols per day, primarily from fruit and coffee, had a 30% lower risk of death from any cause. This was seen in the Italian study, which looked at a large group of people. In addition, a large increase in the chance of the cancer coming back again within five years was seen in the group of patients with stage III colon cancer who followed a high Western diet. Single-cell epigenomic techniques have shown that changes in the diet can affect the methylation levels of immune cells in the tumor microenvironment and make chemotherapy more effective. However, differences in people's genes (for example, variations in a gene called MTHFR C677T) and differences in the bacteria in their intestines can make it hard to predict how well a nutritional treatment will work for someone. This shows that we need personalized nutritional strategies. Studies on animals and people show that diet can have a positive effect on the body. However, there are still significant challenges to implementing this technology in a clinical setting.
References
- Shen, Hui, and Peter W Laird. "Interplay between the cancer genome and epigenome." Cell vol. 153,1 (2013): 38-55. doi:10.1016/j.cell.2013.03.008
- Sun, Linchong et al. "Metabolic reprogramming and epigenetic modifications on the path to cancer." Protein & cell vol. 13,12 (2022): 877-919. doi:10.1007/s13238-021-00846-7
- Dawson, Mark A, and Tony Kouzarides. "Cancer epigenetics: from mechanism to therapy." Cell vol. 150,1 (2012): 12-27. doi:10.1016/j.cell.2012.06.013




