What are SNPs?
Single nucleotide polymorphisms, commonly called SNPs (pronounced “snips”), are the most frequent type of genetic variation among people, accounting for more than 90% of all differences between unrelated individuals. A SNP represents a difference in a single nucleotide. The DNA sequence contains four types of nucleotide bases: A (adenine), C (cytosine), G (cytosine), and T (thymine). For example, SNP variation occurs when an A replaces one of the other three nucleotide bases.
The frequency of less common bases must be at least 1%, otherwise, they are considered as rare mutations. SNPs occur almost once in every 1,000 nucleotides on average, which means a person’s genome contains roughly 4 to 5 million SNPs. Most SNPs are found outside of coding regions. Some SNPs found within a coding region are of particular interest to scientists as they are very likely to alter the biological function of a protein.
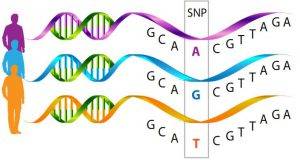
Figure 1. Single nucleotide polymorphism (SNPs).
SNPs are valuable biological markers
Since SNPs occur frequently throughout the genome and tend to be relatively stable genetically, they can serve as excellent biological markers. It means SNPs can be easily tracked, and used for constructing chromosome maps that exhibit the position of known genes and markers, relative to each other. Chromosome maps allow researchers to study traits resulting from the interaction of several genes. The public SNP database (dbSNP) is available for facilitating the detection and cataloging of SNPs. SNP markers have been widely used for forensic purposes.
How to analyze SNPs?
There are many choices for generating whole-genome or small-scale SNP patterns, including SNP microarray, MassARRAY® iPLEX GOLD platform, TaqMan genotyping platform, SNaPshot Multiplex System, and sequencing solutions (such as genotyping by sequencing). In disease studies, researchers first acquire the SNP patterns from individuals affected by the disease and healthy individuals. Next, they compare these patterns (this process are called an “association study”) to identify the most likely associated with the disease-causing genes. Thus, SNP profiles of the disease will be established.
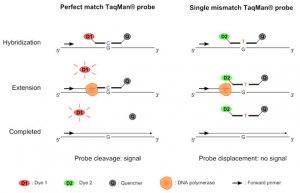
Figure 2. TaqMan SNP genotyping.
Applications of SNP analysis
SNP genotyping has been widely used in a range of fields to detect or target SNP variants for diverse purposes.
- Human health and disease
Most SNPs have no effect on human health or development, while some SNPs have been proven to be associated with human health. Researchers have found that most SNPs serve as molecular markers for pinpointing a disease on the human genome map instead of being responsible for a disease state, since they are commonly located near genes found to be associated with certain diseases. Occasionally, SNPs may cause disease.
SNPs also affect the patients’ response to a drug therapy, which will be helpful in determining and understanding the individual difference in their abilities to absorb and clear certain drugs, as well as predicting adverse side effects to a certain drug. In the future, personalized medicine will help determine the most appropriate drug for an individual by analyzing the patient’s SNP profile, and allow pharmaceutical companies to develop more individualized therapies specific to a patients’ needs.
- Plant and animal breeding
The use of molecular markers has revolutionized the plant/animal genetic analysis which in turn accelerated the implementation of molecular breeding. SNPs have unique advantages in this field due to their genome-wide abundance, high variability, ever-increasing sequence information in public databases, and available SNP genotyping platforms and chemistries. Genomic selection can be implemented in plants or animals with SNP and phenotype data, leading to predicted and improved genetic merit in the offspring.

Leave a Reply