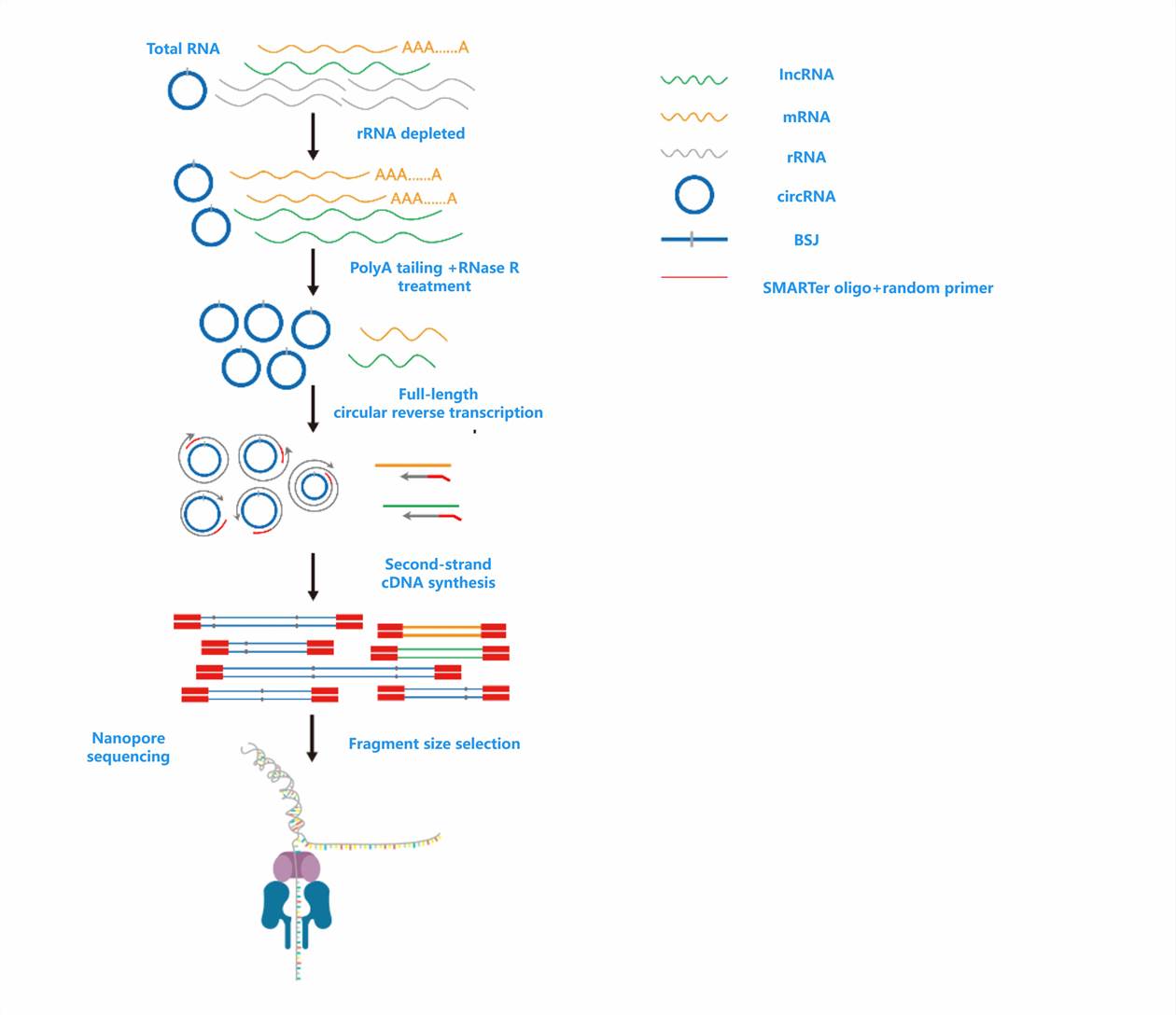
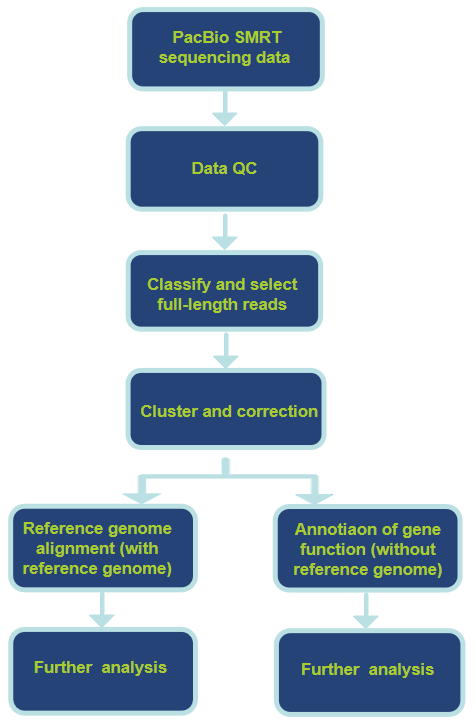
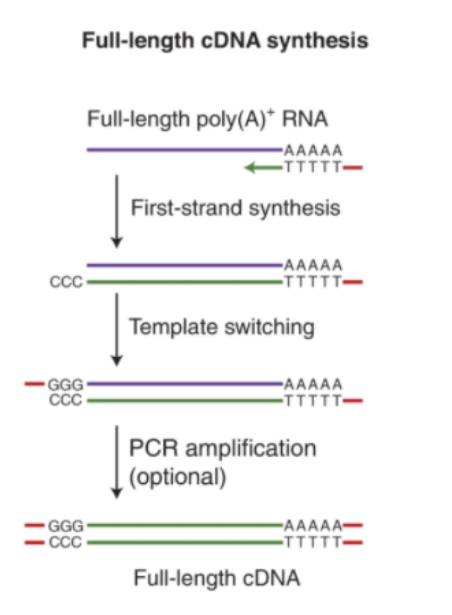
Full-length Transcript & poly(A) Tail Profiling using Nanopore & PacBio
CD Genomics' Poly(A) Tail Length Analysis Service offers expert insight into mRNA 3'-tail dynamics, powered by Nanopore and PacBio platforms. Ideal for biotech professionals, our service delivers:
- Full-length transcript profiling with precise poly(A) tail measurement
- Non-A base detection for U/G/C residues that influence RNA stability
- Isoform-level insights, linking tail features to splicing variants
- Low-input capability, handling down to 0.5 ng RNA—ideal for rare samples
- Comprehensive bioinformatics, including APA site analysis, distribution graphs, and publication-ready visuals
Built for scale and accuracy, our workflow is perfect for transcriptomics, drug development, or crop biotech applications.
- Why Poly(A) Tail Analysis Matters
- Comprehensive Technology Portfolio
- Technology Comparison at a Glance
- Service Workflow
- Key Applications of Poly(A) Tail Length Analysis
- Deliverables
- Demo
- Case Highlight: Deep Dive into Mouse Oocyte Poly(A) Tail Dynamics
Why Poly(A) Tail Analysis Matters
Poly(A) tail length analysis is essential for decoding how mRNA behaves inside cells. These 3' tails:
- Ensure proper mRNA stability and export, keeping transcripts intact long enough to be translated.
- Enhance protein production, by improving ribosome binding during translation.
- Regulate mRNA decay, since shortened tails can signal for degradation.
- Control developmental timing, as seen in embryos where tail length affects gene expression
- Encode epigenetic signals, with non-A residues (U, G, C) influencing function
These mechanisms impact areas like early development, cell stress response, agriculture, and biotech, such as mRNA vaccine design.
Comprehensive Technology Portfolio
CD Genomics provides a complete suite of poly(A) tail length analysis tools, using both legacy and cutting-edge methods on third-generation platforms.
NGS Methods
- TAIL-seq / mTAIL-seq – Ideal for high-throughput poly(A) length and end-modification profiling.
- PAL-seq – Fluorescence-based quantification of poly(A) tails.
- PAT-seq, TED-seq, poly(A)-seq – Targeted studies of specific genes or transcripts.
These techniques offer valuable baseline data and integrate seamlessly into multi-method workflows, supporting our advanced analysis pipeline.
Long-read Sequencing (Our Core Strengths)
Built on Nanopore and PacBio platforms, these methods offer unmatched resolution in poly(A) profiling:
- TAIL Iso-seq (Nanopore)
Captures full-length transcriptome structure, quantifies poly(A) tails, and resolves isoform variants. Widely used in crop research and transcriptomics workflows. Nano 3P-seq (Nanopore)
Ideal for dynamic poly(A) studies. Tracks tail length and internal composition using DOE methods and Nanopore Direct RNA Sequencing.
- FLEP-seq / FLEP-seq2
Advanced isoform-specific tail profiling at genome scale. Developed for simultaneous detection of splicing, APA, and tail length.
- PAIso-seq (PacBio HiFi)
A breakthrough in poly(A) research. Enables full poly(A) tail sequencing with isoform mapping, single-cell sensitivity (≥ 0.5 ng RNA), and detection of U/G/C residues inside tails.
These sophisticated workflows are available as streamlined services, ensuring consistency and reproducibility for your labs.
Technology Comparison at a Glance
| Feature | NGS-based (TAIL/mTAIL/PAL/etc.) | Nanopore Iso-seq | PAIso-seq (PacBio HiFi) |
| Full transcript + poly(A) tail | ❌ | ✅ | ✅ |
| Accurate tail-length measurement | ⚠️ (approximate) | ✅ | ✅ (highest precision) |
| Internal non-A residue detection | ⚠️ | ✅ | ✅ |
| Isoform-level tail mapping | ❌ | ⚠️ partial | ✅ |
| Single-cell / low-input compatible | ❌ | ⚠️ limited | ✅ (0.5 ng sensitivity) |
| Homopolymer sequencing robustness | prone to errors | ✅ | ✅ |
Service Workflow
1. Sample Submission & Quality Control
- Accepts ≥ 0.5 ng total RNA or single-cell lysates for PAIso-seq, and ≥ 100 pg for Nanopore methods.
- Performs RNA quality checks (RIN score, concentration) to ensure reliable downstream results.
2. Library Preparation
- Nanopore platform: TAIL Iso-seq and Nano 3P-seq use oligo(dT) primer capture and template-switching to retain full transcript and tail sequences.
- PacBio platform: PAIso-seq uses SMRTbell library prep optimized for full-length isoforms and poly(A) tracking.
3. Sequencing Execution
- Nanopore: real-time monitoring with fast5 output and basecalling.
- PacBio: high-fidelity (HiFi) long-read mode for precise isoform and tail measurement.
4. Core Bioinformatics
- Tail length called using Nanopolish or Tailfindr for Nanopore reads; proprietary pipelines for PacBio.
- Detects internal non-A residues (U, G, C) in poly(A) bodies.
- Links poly(A) metrics to transcripts, splicing patterns, and alternative polyadenylation (APA) sites.
- Supports differential tail length analysis using tools like NanopLen and linear mixed models (turn0search5).

Key Applications of Poly(A) Tail Length Analysis
Understanding the structure and composition of poly(A) tails enables researchers to uncover hidden layers of post-transcriptional gene regulation. Our platform supports a wide range of research applications across animal, plant, and therapeutic systems.
Embryogenesis & Maternal Transcript Clearance
Understand maternal-to-zygotic transition with isoform-specific tail insights.
Poly(A) tail length and uridylation dynamics are key to controlling maternal mRNA clearance during early development. Our PacBio-based PAIso-seq service has been used to reveal widespread U/G/C incorporation in oocyte transcripts—linking tail length remodeling to developmental stage regulation.
Plant Trait Discovery & Stress Signaling
Map tail-length shifts across tissues, genotypes, or treatments.
TAIL Iso-seq and Nano3P-seq enable poly(A) profiling in roots, leaves, flowers, and pollen. Tail length distributions are tissue-specific and evolutionarily conserved—making them valuable molecular markers for stress response, RNA stability, and gene expression control in crops.
mRNA Therapeutics & Vaccine Design
Optimize translation efficiency by tuning tail length.
Tail composition plays a direct role in mRNA half-life and translation efficiency. Poly(A) profiling can guide rational design of mRNA payloads for vaccines and therapeutics, improving efficacy and consistency in expression.
Single-Cell Transcriptome Tail Profiling
Explore tail-length heterogeneity at single-cell resolution.
Using ultra-sensitive PAIso-seq, we can measure full-length transcripts and poly(A) tails in as little as 0.5 ng of total RNA—ideal for rare cells or limited samples. This enables discovery of isoform-specific regulation even in individual cells.
Alternative Polyadenylation (APA) & Isoform Regulation
Link 3'UTR variation with tail length to decode regulatory programs.
APA influences transcript stability and localization. Our full-length sequencing methods link APA site usage to tail length, enabling discovery of isoform-specific expression patterns under different conditions.
Deliverables
- Raw data: FASTQ/BAM (Nanopore) or HiFi reads (PacBio).
- QC report: sequencing metrics and library statistics.
- Poly(A) analytics: tail-length distribution, non-A residue maps, isoform associations, APA effects.
- Visual output: publication-ready plots, sashimi diagrams (from Nano/PAIso reads), tables for analysis.
- Consultation call: expert review to discuss results and next steps.
Demo

Q1: Which sequencing platforms support this service?
A: We use Oxford Nanopore and PacBio SMRT systems. These long-read platforms enable full-length transcript and poly(A) tail analysis.
Q2: How is poly(A) length measured?
A: On Nanopore, we use tools like nanopolish or tailfindr to estimate tail lengths from signal dwell time in fast5 files. On PacBio, we leverage HiFi read analysis optimized for long homopolymers.
Q3: Can the service detect internal U, G, or C residues?
A: Yes. We process full-length reads to identify internal non-A bases (U/G/C) within poly(A) tails. This adds epigenetic insight to your profiling.
Q4: What analysis workflows are available?
A: Our pipeline delivers:
- Tail-length distribution (histograms, statistics)
- Internal base composition (residue maps)
- Isoform and APA mapping via full-length transcript alignment
- Differential tail-length comparison across samples using tools like NanopLen
Q5: Do long reads reduce accuracy due to homopolymers?
A: Modern algorithms account for homopolymer challenges. Our long-read callers—tailfindr, nanopolish, Nano3P—provide ~88–95% correspondence with known standards.
Q6: Can you analyze APA sites in the same run?
A: Yes. Since we sequence full-length transcripts, we map APA site variation (3' UTR length changes) alongside tail features.
Q7: How are low-input samples handled?
A: We support single-cell or low-input workflows, detecting tails from as little as 0.5 ng RNA (PacBio) and 100 pg (Nanopore).
Q8: How do Nanopore and PacBio results differ?
| Feature | Nanopore | PacBio SMRT HiFi |
| Real-time data | ✅ fast5 + fastq during run | ⚠️ post-run HiFi reads |
| Homopolymer accuracy | ✅ good with tailfindr/nanopolish | ✅ high precision (>200 nt) |
| Internal base detection | ✅ yes | ✅ yes |
| Isoform-level resolution | ✅ full-length reads | ✅ full-length reads |
Q9: Which tool best suits my project—Nanopore or PacBio?
A:
- Use Nanopore for fast turnaround, flexible long-read requirements, and real-time monitoring.
- Choose PacBio when you need higher accuracy, longer reads, and sensitive detection in low-input or single-cell samples.
Q10: What's the minimum RNA input?
A: We accept as little as 0.5 ng total RNA for PAIso-seq and ≥ 100 pg for Nanopore workflows.
Q11: Can you detect U/G/C residues in the poly(A) tail?
A: Yes. Our Nanopore and PacBio pipelines are validated to map internal non-A residues.
Q12: Is this service suitable for mRNA vaccine research?
A: Absolutely. Designed for non-clinical R&D, our workflow supports GMP-ready mRNA design, optimizing tail length and composition for expression outcomes.
Case Highlight: Deep Dive into Mouse Oocyte Poly(A) Tail Dynamics
Study Context
A landmark study by Lu et al. (2019) introduced PAIso-seq, a highly sensitive method for full-length transcript profiling paired with poly(A) tail analysis. Remarkably, the technique required only 0.5 ng of total RNA from a single mouse germinal vesicle (GV) oocyte—an extremely limited, yet biologically rich sample.
What They Discovered
Widespread internal non-A residues
‣ Analysis showed 17% of mRNAs contained U, G, or C within their poly(A) tails (not just at the tail end) .
Isoform-specific tail variations
‣ Same-gene transcripts exhibited differing tail lengths. For instance, Ccnb1 isoforms had distinct poly(A) profiles.
Biological relevance
‣ Transcripts with longer tails correlated with higher protein levels, linking tail length to translational control in maturing oocytes .

The principle and validation of PAIso-seq
Why This Matters
- textbook views: It revealed poly(A) tails are not uniform A-strings; their composition varies in-cell and among isoforms.
- Enables precision biology: PAIso-seq unlocked tail-level insight at single-cell resolution in rare samples.
- Advances tool development: The approach, including full-length capture and high-fidelity sequencing, became a reference for later studies like PAIso-seq2, which further explored tail remodeling with and without polyadenylation .
References
- Liu, Y., Nie, H., Liu, H. et al. Poly(A) inclusive RNA isoform sequencing (PAIso-seq) reveals wide-spread non-adenosine residues within RNA poly(A) tails. Nat Commun 10, 5292 (2019). https://doi.org/10.1038/s41467-019-13228-9
- Jia J, Lu W, Liu B, Fang H, Yu Y, Mo W, Zhang H, Jin X, Shu Y, Long Y, Pei Y, Zhai J. An atlas of plant full-length RNA reveals tissue-specific and monocots-dicots conserved regulation of poly(A) tail length. Nat Plants. 2022 Sep;8(9):1118-1126. DOI: 10.1038/s41477-022-01224-9
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment