HiChIP Service: Unlock 3D Genome Interactions with Fast, Low-Input, High-Resolution
At CD Genomics, we bring years of hands-on experience in HiChIP assay development and optimisation. Our team has successfully delivered numerous high-quality HiChIP datasets for clients across academia and industry.
- Proven Expertise: Extensive track record in performing complex HiChIP experiments from diverse cell types.
- Data Quality Assurance: Each project is guided by stringent QC metrics to ensure reliable, publication-ready results.
- End-to-End Support: From experimental design to advanced bioinformatics analysis, we provide full-service solutions tailored to your research goals.
At a glance
- Workflow Overview
- Key Applications of HiChIP
- Advantages of HiChIP
- Sequencing Specifications
- HiChIP Sample Requirements
- Demo Results
- Frequently Asked Questions
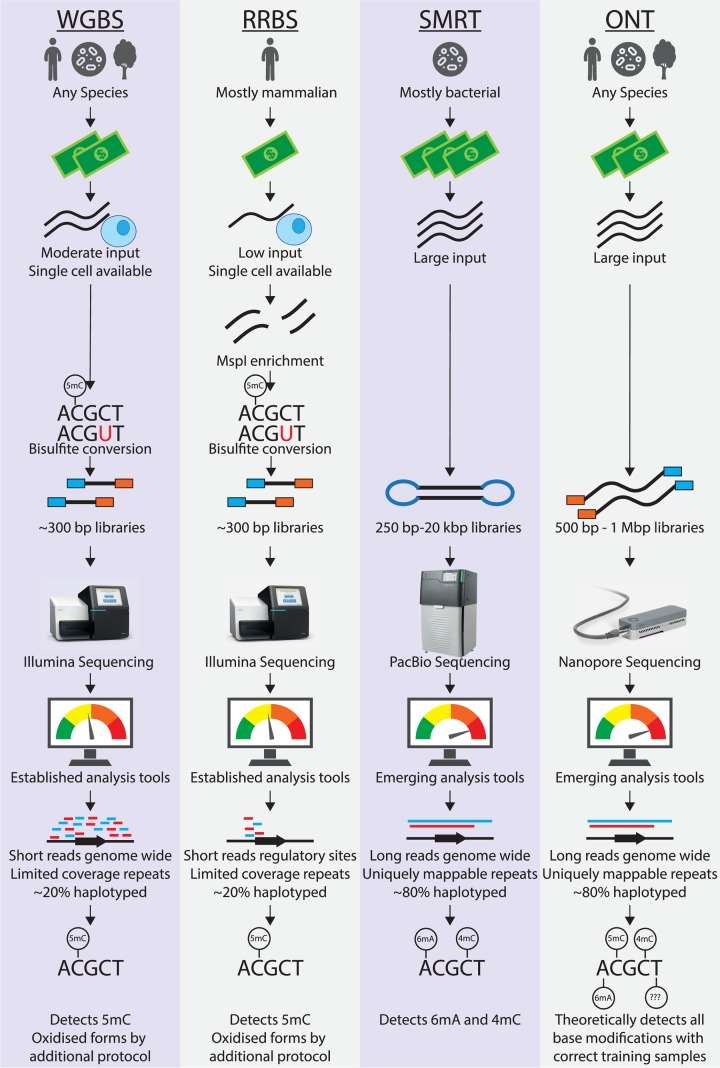
What Is HiChIP?
HiChIP is an advanced genomic technique used to map protein-directed DNA-DNA interactions within the 3D architecture of the nucleus. By integrating the in situ chromatin interaction capture strategy of Hi-C with the specificity of chromatin immunoprecipitation (ChIP), HiChIP enables researchers to map high-resolution chromatin contacts centered on specific DNA-binding proteins.
This hybrid approach delivers three major advantages:
- High sensitivity for detecting protein-mediated chromatin loops
- Greater resolution compared to traditional Hi-C
- Minimal input requirements—ideal for limited or precious cell samples
Unlike ChIA-PET, which enriches target-bound DNA before spatial interaction capture, HiChIP reverses the order. It starts with in situ chromatin crosslinking and ligation, followed by targeted enrichment of protein-associated complexes via ChIP. This workflow improves efficiency and reduces background noise.
How HiChIP Works
The HiChIP protocol integrates steps from both Hi-C and ChIP-seq, optimising them for protein-centric chromatin interaction studies:
In situ crosslinking: Cells are fixed to preserve chromatin structure.
Spatial interaction capture: Chromatin is fragmented and in situ linkage is performed to preserve interactions between adjacent genomic regions.
ChIP enrichment: An antibody targeting the protein of interest pulls down DNA-protein complexes.
DNA purification and sequencing: Crosslinks are reversed, and biotin-labeled ligation products are purified and sequenced to reveal genome-wide interaction maps.
 Overview of the HiChIP experimental principle.
Overview of the HiChIP experimental principle.
HiChIP is particularly valuable for studying how transcription factors, histone modifications, or chromatin remodelers influence gene regulation across topologically associated domains (TADs).
Why Use HiChIP?
Researchers exploring 3D genome organisation often run into limitations when relying on single-method approaches like Hi-C or ChIP-seq. That's where HiChIP comes in—a hybrid technique that answers both the "where" and the "how" of protein-DNA interactions.
The Gaps in Traditional Methods
- Hi-C can chart genome-wide chromatin loops, but it's like getting a city map without landmarks. You see the connections—but not who built them.
- ChIP-seq shows where specific proteins bind on DNA, but offers a flat, one-dimensional view. It lacks the spatial insight needed to understand how these sites interact across the genome.
The HiChIP Breakthrough
HiChIP merges the strengths of both methods into a single, protein-targeted chromatin interaction tool.It pinpoints where a protein binds (like ChIP-seq).
It also maps which distant genomic regions physically connect at those sites (like Hi-C).
This dual capacity makes HiChIP especially powerful for:
- Investigating enhancer-promoter loops
- Understanding chromatin architecture, such as DNA loops and domains
- Uncovering the spatial gene regulation behind complex diseases
For teams focused on functional genomics or disease pathway analysis, HiChIP is a game-changer—offering clarity where older methods fall short.
Comparison of ChIA-PET and HiChIP Technologies
| Feature | ChIA-PET | HiChIP |
| Principle | Chromatin immunoprecipitation (ChIP) is performed first to enrich DNA bound to the target protein, followed by spatial chromatin interaction linking. | Begins with in situ crosslinking and spatial chromatin interaction linking, then performs ChIP, followed by Tn5 transposase-based library construction. |
| Sensitivity | Moderate | High |
| Sample Input Requirement | High | Low |
| Advantages | High resolution; enriches DNA interactions directed by the target protein. | Lower cost; high sensitivity; compatible with low-input samples. |
| Limitations | Requires complex library prep and high input; difficult and costly to optimise. | May introduce bias during immunoprecipitation or ligation; requires antibody optimisation. |
| Best Use Case | Ideal for well-funded projects focusing on large-scale, comprehensive profiling of protein-guided chromatin interactions. | Suited for low-input or large-scale profiling of transcription factor– and histone-modification–driven chromatin architecture in functional genomics or drug discovery. |
Workflow Overview
HiChIP follows a streamlined, in situ protocol to capture protein-directed chromatin interactions:
Crosslinking
Cells or tissues (animal or plant) are treated with formaldehyde to stabilise chromatin structures.
Chromatin Fragmentation & Spatial Interaction Linking
The genome is enzymatically fragmented, and spatially adjacent DNA fragments are ligated.
Immunoprecipitation (ChIP)
Antibodies are used to enrich for DNA fragments interacting with the target protein.
Library Construction & Sequencing
Chimeric DNA fragments are purified, indexed, and sequenced to identify genome-wide interaction patterns.
 Simplified workflow of HiChIP sequencing service
Simplified workflow of HiChIP sequencing service
Key Applications of HiChIP
Chromatin Interaction Profiling for Specific Proteins
HiChIP enables detailed mapping of where a protein binds and what distal regions it interacts with—ideal for studying chromatin architecture around specific regulators.
Mechanistic Studies of Transcription Factors
By identifying transcription factor binding sites across the genome, HiChIP helps clarify how these proteins control gene expression networks.
Epigenetic Regulation Analysis
HiChIP can uncover how histone modifications (e.g. methylation, acetylation) influence gene activity through long-range DNA contacts.
Dynamic Chromatin Architecture Studies
The method captures changes in chromatin folding across time or conditions, providing insight into dynamic 3D genome reorganisation.
HiChIP is increasingly used in functional genomics to understand how spatial genome organisation underpins gene regulation and cell identity.
Advantages of HiChIP
1. Exceptional Sensitivity with Low Input Requirements
Traditional Hi-C typically requires millions of cells to generate high-resolution contact maps. HiChIP overcomes this barrier with antibody-based enrichment, reducing input needs to just 100,000–500,000 cells.
Li et al. (2019) used only 50,000 human embryonic stem cells to map OCT4-driven regulatory networks in pluripotency.
2. Targeted and Efficient Data Output
Unlike Hi-C, which captures genome-wide interactions, HiChIP selectively retains only those contacts associated with the target protein. This reduces data volume by up to 90%, streamlining downstream analysis.
Sun et al. (2017) demonstrated how HiChIP pinpoints CTCF-bound boundaries of topologically associated domains (TADs), revealing its role as a "genome architect."
3. Dynamic Regulation Across Time or Condition
HiChIP is ideal for comparing chromatin interactions across developmental stages or disease states, offering fine time-resolution insights.
Zhang et al. (2022) showed that NF-κB rapidly remodels the 3D interaction landscape of inflammation-related genes during immune activation.
4. Multi-Omics Integration for Deeper Insights
HiChIP data can be integrated with ATAC-Seq and RNA-Seq to build a holistic view of how genome structure influences function and expression.
Cipta NO et al. (2025) used this approach to construct a comprehensive model linking chromatin structure, gene accessibility, and transcriptional output.
Sequencing Specifications
· Platform: Illumina NovaSeq 6000
· Read Mode: Paired-End 150 bp (PE150)
· Recommended Data Yield: 90–100 Gb per sample
HiChIP Sample Requirements
| Category | Requirement |
| Sample Type | - Fresh live cells - Formaldehyde-crosslinked cells (preferred) |
| Minimum Input | ≥ 1 × 107 crosslinked cells per sample |
| Accepted Species | Human, Mouse, Rat |
| Other Species | Require pre-evaluation before sample submission |
HiChIP Bioinformatics Analysis Overview
| Content | Description |
| 1. Read Alignment | Align raw sequencing reads to the reference genome |
| 2. Quality Control | Evaluate experiment quality (e.g., alignment stats, valid interaction pairs) |
| 3. Interaction Heatmaps | Generate contact maps at multiple resolutions |
| 4. Loop Detection | Identify chromatin loops based on spatial interaction signals. |
| 5. Loop Annotation | Map loops to genomic features (e.g., enhancer–promoter connections) |
| 6. Hub/Super-Enhancer Detection | Identify high-density interaction hubs and super-enhancer-associated loops |
| 7. Data Visualization Files | Provide track files for genome browser display (e.g., .hic, .bedpe, .bigWig) |
| 8. Differential Loop Analysis | Compare loop structures between conditions or sample groups |
| 9. GO & KEGG Pathway Analysis | Functional enrichment of genes near significant chromatin interactions |
Demo Results
 Summary of key HiChIP data analysis outputs.
Summary of key HiChIP data analysis outputs.
Includes a loop distance distribution histogram, alignment and duplication statistics, FDR-based significant interaction count, and a loop MA plot highlighting differential chromatin interactions.
 Visual summary of HiChIP differential interaction analysis.
Visual summary of HiChIP differential interaction analysis.
Includes a volcano plot showing differentially enriched loops, a contact decay curve across genomic distances, and both box plot and violin plot visualizations of loop intensities between conditions.
Frequently Asked Questions
What is the difference between HiChIP and ChIA-PET?
HiChIP employs an in situ chromatin adjacency linking step before ChIP enrichment and Tn5-based library construction. In contrast, ChIA PET conducts ChIP enrichment first before fragment linkage, often demanding millions of cells and deeper sequencing. HiChIP achieves comparable resolution with significantly reduced input and cost.
How low can cell input be for HiChIP?
Our optimized HiChIP workflow delivers high-quality data with as few as 50,000 to 100,000 crosslinked cells. That's over 100× less than ChIA-PET or standard Hi-C methods, making HiChIP ideal for rare or sensitive samples .
Does HiChIP work on plant or non-model species?
While most published HiChIP studies focus on human and mouse, the protocol is adaptable. For non-model organisms, we evaluate crosslinking efficiency, enzyme compatibility, and antibody specificity on a case-by-case basis to ensure success in custom applications.
How should I prepare cells for HiChIP sequencing?
We recommend submitting formaldehyde-crosslinked cells (≥ 1×107 per sample) or fresh live cells. Proper fixation and cell integrity are critical. Detailed sample prep SOPs and QC guidelines are provided to help ensure successful experimental outcomes.
What QC metrics are used in HiChIP library assessment?
We evaluate both quantitative and qualitative metrics:
- No-duplication read pairs: Reflects alignment quality and library complexity (higher is better)
- Valid interaction pairs: High percentage of reads mapped within expected restriction-enzyme fragment ranges
- Cis/trans interaction ratios and enrichment near binding peaks: Confirms proximity ligation specificity
These metrics help decide whether to proceed to high-depth sequencing or adjust protocol steps.
How does HiChIP overcome traditional Hi-C or ChIP-seq limitations?
HiChIP integrates in situ chromatin adjacency linking with ChIP enrichment, providing targeted, protein centric 3D interaction maps with greatly reduced input and sequencing requirements.
What types of targets and proteins work well with HiChIP?
Ideal targets include transcription factors, histone marks (e.g. H3K27ac, H3K4me3), or architectural proteins like CTCF. We offer antibody consultation to ensure target compatibility
What bioinformatics deliverables can I expect?
We provide fully processed outputs including loop detection, annotation (e.g., enhancer–promoter), heatmaps, differential analysis, and functional enrichment (GO/KEGG). Browser files like .bedpe and bigWig are included
Can HiChIP data be integrated with other omics like RNA-Seq or ATAC-Seq?
Absolutely. Integrating HiChIP with ATAC-Seq or RNA-Seq uncovers how 3D chromatin structure links to DNA accessibility and gene expression for comprehensive regulatory insights
References
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, Chang HY. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016 Nov;13(11):919-922. doi: 10.1038/nmeth.3999. Epub 2016 Sep 19. PMID: 27643841; PMCID: PMC5501173.
- Ramirez RN, Chowdhary K, Leon J, Mathis D, Benoist C. FoxP3 associates with enhancer-promoter loops to regulate Treg-specific gene expression. Sci Immunol. 2022 Jan 14;7(67):eabj9836. doi: 10.1126/sciimmunol.abj9836. Epub 2022 Jan 14. PMID: 35030035; PMCID: PMC9059705.
- Giambartolomei C, Seo JH, Schwarz T, Freund MK, Johnson RD, Spsiak S, Baca SC, Gusev A, Mancuso N, Pasaniuc B, Freedman ML. H3K27ac HiChIP in prostate cell lines identifies risk genes for prostate cancer susceptibility. Am J Hum Genet. 2021 Dec 2;108(12):2284-2300. doi: 10.1016/j.ajhg.2021.11.007. Epub 2021 Nov 24. PMID: 34822763; PMCID: PMC8715276.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment