Long-read ChIA-PET Sequencing Service for 3D Genome Mapping
At CD Genomics, we offer end-to-end ChIA-PET services tailored for 3D genome research. Our service includes:
- Library construction
- Sequencing
- Data analysis
With extensive hands-on experience in 3D genomics, our team has generated a wide range of high-quality ChIA-PET datasets, supporting reliable, publication-ready results for transcriptional regulation and chromatin architecture studies.
At a glance:
- What is ChIA-PET
- Advantages and Limitations of ChIA-PET Technology
- Sequencing Specifications
- How ChIA-PET Works: Step by Step
- Applications of ChIA-PET Technology in 3D Genome Research
- Bioinfomatics Analysis
- ChIA-PET Sample Requirements
- Demo Results
- Frequently Asked Questions
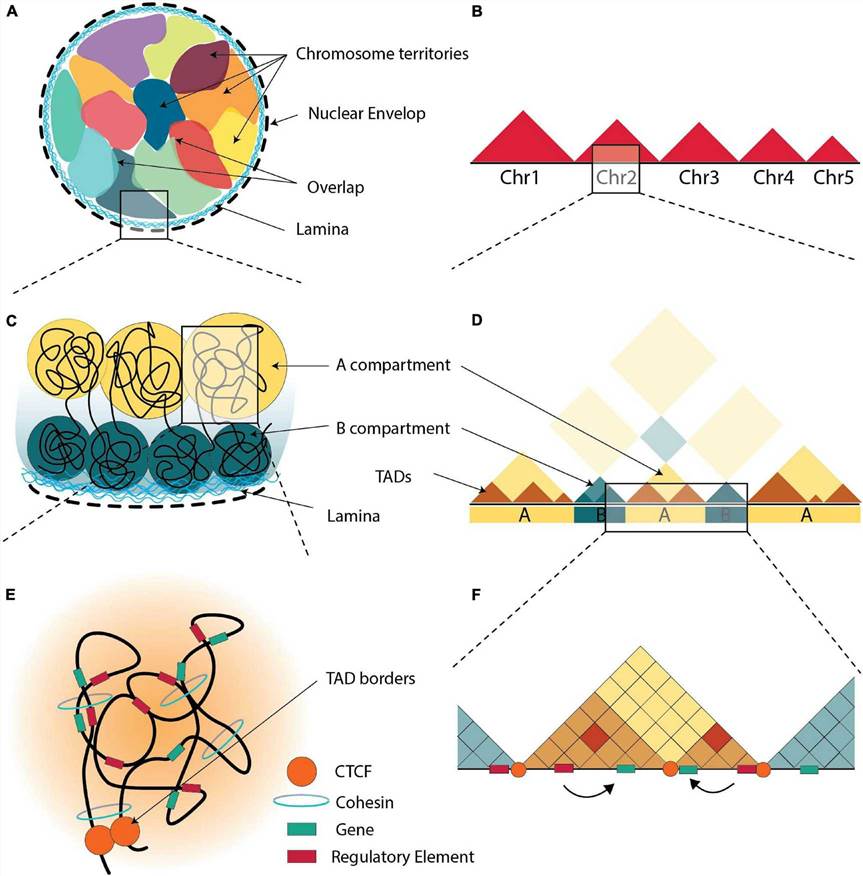
What is ChIA-PET
ChIA-PET (Chromatin Interaction Analysis by Paired-End Tag sequencing) is an advanced method designed to chart protein-mediated DNA interactions across the entire genome. It excels at identifying long-range regulatory loops—such as promoter–promoter and promoter–enhancer interactions—that play a crucial role in gene expression.
By integrating chromatin immunoprecipitation (ChIP), spatial adjacency-mediated fragment linking, paired-end tag (PET) sequencing, and long-read sequencing (NGS), ChIA-PET maps the three-dimensional genome architecture. This integrated approach enables investigation of how chromatin folding and looping affect gene regulation.
Unlike traditional ChIP-seq, which only detects linear binding sites of transcription factors, ChIA-PET goes further by mapping physical interactions between distant genomic regions bound by the same protein of interest. For example, in breast cancer cells, ChIA-PET has been used to define interaction networks among estrogen receptor α (ERα) binding sites—offering insights into hormone-driven gene regulation.
As more protein factors and corresponding antibodies become available, ChIA-PET can be applied to investigate genome-wide interactions related to transcriptional regulation, DNA replication, and chromatin remodeling. These insights are key to understanding how genes are turned on and off—and how misregulation can lead to disease.
For researchers aiming to decode the dynamic architecture of the genome, ChIA-PET offers a high-throughput, data-rich solution for visualizing regulatory complexity in situ.
 Schematic diagram illustrating the principle of ChIA-PET, showing key steps from chromatin fragmentation to formation of paired-end tags (PETs) for interaction mapping.
Schematic diagram illustrating the principle of ChIA-PET, showing key steps from chromatin fragmentation to formation of paired-end tags (PETs) for interaction mapping.
Advantages and Limitations of ChIA-PET Technology
ChIA-PET is a genome-wide, unbiased, de novo method designed to capture chromatin interactions mediated by protein–DNA binding. It offers a powerful combination of specificity and spatial resolution—especially for researchers focused on 3D genome architecture and gene regulation.
Key Strengths of ChIA-PET
Dual-level insight
ChIA-PET identifies both the protein binding sites and the interactions between those sites within the same experiment—something most conventional techniques cannot achieve.
Minimizes noise from random ligation
Instead of using restriction enzymes that may introduce random chromatin contacts, ChIA-PET applies sonication to fragment protein–DNA complexes, significantly reducing background noise in interaction data.
Combines ChIP specificity with 3D interaction mapping
By using protein-specific antibodies, ChIA-PET retains the targeting precision of ChIP-seq, while extending its utility to uncover long-range chromatin interactions—without the added complexity of whole-genome 3D profiling.
Comparison of ChIP-seq, ChIP-PET, and ChIA-PET
| Technique | Type | Application Focus | Key Features |
| ChIP-seq (single-end) | Protein–DNA interaction | Maps transcription factor binding | Reads only a short region (~5' end); average fragment length ~500 bp; limited spatial resolution |
| ChIP–PET (paired-end) | Protein–DNA interaction | Captures both ends of DNA fragments | Useful for detecting fusion transcripts, chromosomal rearrangements, and molecular interactions |
| ChIA–PET (paired-end) | Chromatin–chromatin interaction | 3D genome research | Links binding site detection with spatial interaction mapping in one assay; ideal for uncovering regulatory loops |
When to Use ChIA-PET Over Other Methods
If your research requires both high-resolution transcription factor binding data and insight into long-range DNA interactions—especially in the context of enhancer–promoter looping—ChIA-PET is the most comprehensive solution.
Sequencing Specifications
| Specification | Details |
| Read format | Paired-end (e.g. 2 × 150 bp or 2 × 250 bp) |
| Sequencing depth | ~200–300 million raw read pairs |
| Expected PET output | ~40 million unique, non-redundant PETs |
| Library design | Bridge-linker or half-linker protocol with MmeI enzyme |
| QC metrics | ≥10M PETs, intra/inter PET ratio ≥ 1, high mapping rate, low duplication |
| Processing pipeline | ChIA-PIPE or ChIA-PET2 recommended or ChIA-PET Tool V3 |
How ChIA-PET Works: Step by Step
ChIA-PET Workflow: Step-by-Step Protocol for Capturing DNA Interactions
a. Crosslinking Chromatin Complexes
Tissues are first treated with formaldehyde and EGS to crosslink proteins and stabilize all endogenous DNA–DNA interactions mediated by protein factors.
b. Immunoprecipitation of Protein–DNA Complexes
Target-specific antibodies are used to enrich for DNA–protein complexes that involve the protein of interest, capturing relevant chromatin regions.
c. DNA End Processing and Ligation
After purification, DNA ends within the complex are repaired. Specialized adaptors are then ligated to the two ends of each DNA molecule to form a single, circularized DNA tag.
d. Reverse Crosslinking and DNA Fragmentation
Crosslinks are reversed, and the DNA is digested to release ligated complexes. These hybrid DNA molecules (formed from ligated ends) are further purified and used to build a sequencing library.
e. Sequencing and Data Analysis
High-throughput sequencing is performed to map interaction sites across the genome, allowing for 3D reconstruction of regulatory DNA architecture.
 ChIA-PET sequencing service workflow, from crosslinking and immunoprecipitation to linker ligation, inverse PCR, and paired-end sequencing.
ChIA-PET sequencing service workflow, from crosslinking and immunoprecipitation to linker ligation, inverse PCR, and paired-end sequencing.
Applications of ChIA-PET Technology in 3D Genome Research
Mapping long-range DNA regulatory interactions
ChIA-PET identifies genome-wide chromatin interactions mediated by transcription factors or modified histones, such as promoter–promoter and promoter–enhancer loops.
Constructing high-resolution 3D genome architecture maps
The technology enables researchers to visualize how regulatory DNA elements are spatially organized within the nucleus, supporting functional annotation of non-coding regions.
Integrating with transcriptomic data for regulatory insights
When combined with RNA-seq or GRO-seq, ChIA-PET helps uncover how distant cis-regulatory elements work together to control transcription.
Providing actionable targets for genome editing
By revealing protein-mediated DNA interactions that drive gene expression, ChIA-PET can guide CRISPR/Cas9 targeting and functional validation studies.
Bioinfomatics Analysis
| Category | Service Content |
| Raw Data Processing | Adapter/linker trimming, quality filtering, and removal of low-quality or unmapped reads. |
| Genome Mapping | Accurate alignment of paired-end tags (PETs) to the reference genome using tools like BWA. |
| Interaction Pairing | Reconstruction of valid PETs and filtering of artifacts such as self-ligation or dangling ends. |
| Deduplication | Removal of PCR duplicates to improve interaction confidence and downstream accuracy. |
| Peak Calling | Identification of protein-binding regions from enriched PET clusters, similar to ChIP-seq. |
| Loop Detection | Detection of chromatin loops formed between binding regions (e.g., enhancer–promoter). |
| Significant Loop Calling | Statistical modeling to identify high-confidence loops based on background and noise estimation. |
| Quality Control | Generation of comprehensive QC metrics: PET yield, mapping rate, duplication rate, interaction types. |
| Visualization Outputs | Generation of loop tracks, peak BED files, and contact matrices for genome browsers or 3D genome viewers. |
| Optional Analysis | Allele-specific loop analysis using phased genotype data to uncover cis-regulatory variation. |

ChIA-PET Sample Requirements
| Category | Requirement |
| Sample Type | − Fresh live cells or ≥ 1% formaldehyde-crosslinked cells |
| Minimum Input | − ≥ 10 million crosslinked cells per sample<br>− In situ protocol: as few as 1 million cells |
| Accepted Species | − Human, Mouse, Rat |
| Other Species | − Require pre-evaluation before sample submission |
| Target Protein | − Specify target protein (e.g. CTCF, RNAPII) and validated antibody (vendor, clone, validation) |
Demo Results
 ChIA-PET demo result panel showcasing six key visual outputs: loop track, peak track, contact matrix, APA plot, arc plot, and allele-specific loop analysis—demonstrating 3D chromatin interaction insights from sequencing data.
ChIA-PET demo result panel showcasing six key visual outputs: loop track, peak track, contact matrix, APA plot, arc plot, and allele-specific loop analysis—demonstrating 3D chromatin interaction insights from sequencing data.
Frequently Asked Questions
1. What is ChIA-PET sequencing, and why use it?
ChIA-PET (Chromatin Interaction Analysis by Paired-End Tag sequencing) combines ChIP and PET sequencing to map long-range chromatin interactions across the genome. It identifies both protein binding sites and their interactions, enabling a 3D view of gene regulation networks.
2. What sample type do I need to send?
We accept fresh live cells or ≥ 1% formaldehyde-crosslinked cells. Adhering to dual fixation (formaldehyde + EGS) improves recovery of protein-mediated DNA loops .
3. How many cells are required per sample?
Standard ChIA-PET requires ≥ 10 million crosslinked cells.
If doing an in-situ variant, 1 million cells may be sufficient, depending on antibody quality and target abundance.
4. Which species are supported?
We routinely support human, mouse, and rat samples. Other species require prior evaluation. Please contact us before submission.
5. What antibody information must I provide?
Please supply antibody clone, vendor, host species, and validation evidence (e.g. ChIP seq or Western blot). A validated antibody ensures high signal-to-noise and reproducible results.
6. How much sequencing depth do I need?
We recommend 200–300 million paired-end reads per sample. This often yields ~40 million unique non-redundant PETs needed to detect statistically significant chromatin loops .
7. What quality metrics do you report?
- Unique PET count (target ≥ 10 million after filtering)
- Intra-chromosomal/inter-chromosomal PET ratio (ideally ≥ 1, preferably >2)
- Mapping rate (≥ 70%), duplication rate, and clustering metrics.
8. What output files will I receive?
Standard deliverables include:
- Raw FASTQ files (R1 & R2)
- QC report with all metrics
- Optional: binding peak calls, chromatin loop files, and contact matrices for visualization
9. What are ChIA-PET's advantages over HiChIP or PLAC-seq?
ChIA-PET uses ChIP before ligation, ensuring only interactions mediated by the target protein are captured. This provides higher specificity but requires more input material and sequencing depth compared to HiChIP or PLAC-seq
References
- 1.Nakato R, Sakata T. Methods for ChIP-seq analysis: A practical workflow andadvanced applications. Methods. 2021;187:44-53. doi:10.1016/j.ymeth.2020.03.005.
- 2.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10(10):669-680. doi:10.1038/nrg2641.
- 3.Kaya-Okur HS, Wu SJ, Codomo CA, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10(1):1930. Published 2019 Apr 29. doi:10.1038/s41467-019-09982-5.
- 4.Yue Q, Wang Z, Shen Y, et al. Histone H3K9 Lactylation Confers Temozolomide Resistance in Glioblastoma via LUC7L2-Mediated MLH1 Intron Retention. Adv Sci (Weinh). 2024;11(19):e2309290. doi:10.1002/advs.202309290.
- 5.Li X, Luo OJ, Wang P, et al. Long-read ChIA-PET for base-pair-resolution mapping of haplotype-specific chromatin interactions. Nat Protoc. 2017;12(5):899-915. doi:10.1038/nprot.2017.012.
- 6.Mumbach MR, Rubin AJ, Flynn RA, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13(11):919-922. doi:10.1038/nmeth.3999.
- 7.Ramirez RN, Chowdhary K, Leon J, Mathis D, Benoist C. FoxP3 associates with enhancer-promoter loops to regulate Treg-specific gene expression. Sci Immunol. 2022;7(67):eabj9836. doi:10.1126/sciimmunol.abj9836.
- 8.Giambartolomei C, Seo JH, Schwarz T, et al. H3K27ac HiChIP in prostate celllines identifies risk genes for prostate cancer susceptibility. Am J Hum Genet. 2021;108(12):2284-2300. doi:10.1016/j.ajhg.2021.11.007.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment