Functional metagenomics is a powerful method for discovering new biocatalysts from a wide range of environmental samples. The building of metagenomic libraries is usually accompanied by the selection of clones expressing the desired phenotype, such as enzymatic or antimicrobial activity, antibiotic/xenobiotic resistance, or the capacity to disintegrate toxic compounds or environmental pollutants. Gene annotation is based on phenotypic based screening of metagenomic clones, which provides invaluable information about gene function. This approach has recently been used to successfully identify amylolytic, cellulolytic, and lipolytic activities with potential industrial applications. Antimicrobial bioprospecting and bioremediation of soils contaminated with aromatic compounds can also benefit from functional metagenomics. The functional metagenomics approach is used in ARG research to select clones on agar plates supplemented with antibiotics, then analyze their genetic content.
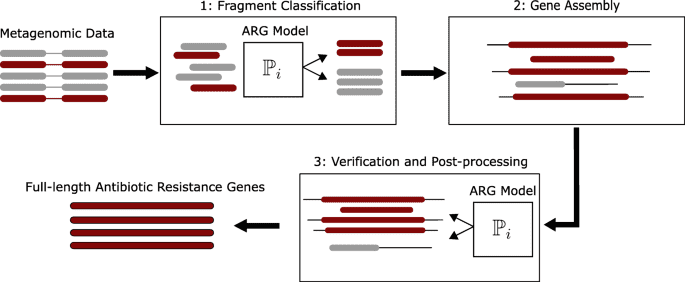
Figure 1. Identification and reconstruction of novel antibiotic resistance genes from metagenomes. (Berglund, 2019)
Metagenomic Analysis of Resistance Genes
Antibiotic resistance research has been focused on pathogenic bacteria for several years. This line of investigation has aided in the discovery of key mechanisms that mediate bacterial antibiotic resistance. The four major processes of antibiotic resistance are: I enzymatic modification or destruction of the antibiotic, which typically involves the overproduction of enzymes that neutralize the antibiotic, (ii) modification of the antibiotic target molecule to lessen its binding capacity, and (iii) alteration of metabolic pathways and regulatory networks to avoid the antibiotic’s effect. and (iv) reduction of the intracellular accumulation of the antibiotic by lessening cellular permeability to it or activating efflux mechanisms to export the harmful molecule.
Essential advances in our knowledge of bacterial resistance, genes that provides resistance, and other phenotypes of interest have been made thanks to mass sequencing and complete genome analysis. Furthermore, genomic data has revealed bacteria’s remarkable genetic plasticity, which allows them to react to a wide range of threats, including antibiotics. However, metagenomic methods are progressively being used to comprehend the functioning of sets of genes that can obtain antibiotic resistance in resistomes. Function-based or sequence-based metagenomic approaches are both possible. Multiple sequence reads are generated and analyzed using sequence analysis methods.
Complete genome sequencing is the most proactive solution for metagenome sequencing; it allows researchers to investigate the structural and functional diversity of a microbial community by identifying genes and metabolic pathways and reconstructing nearly complete bacterial genomes. The main benefit of this method is its sensitivity, which allows for the detection of more species and the identification of potential ARGs.
Applications
Although antibiotic resistance in pathogenic and clinically important bacteria has been extensively studied, little is known about antibiotic resistance genes’ environmental diversity (ARGs). Even before they came to our attention and revolutionized medicine in the antibiotic era, ARGs and antibiotics were two aspects of life for microorganisms in the environment. Metagenomics and other culture-independent techniques are now revealing the diversity of antibiotic/antibiotic resistance systems found in the environment. Acknowledging antibiotic resistance mechanisms could lead to the discovery of new drug targets and the development of new and efficient antibiotic molecules. In the fight against microbial diseases, studying ARGs is a welcome strategy.
References:
- de Abreu VA, Perdigão J, Almeida S. Metagenomic Approaches to Analyze Antimicrobial Resistance: An Overview. Frontiers in Genetics. 2020;11.
- Berglund F, Österlund T, Boulund F, et al. Identification and reconstruction of novel antibiotic resistance genes from metagenomes. Microbiome. 2019 Dec;7(1).
- Gião MS, Keevil CW. Listeria monocytogenes can form biofilms in tap water and enter into the viable but non-cultivable state. Microbial ecology. 2014 Apr;67(3).
- Garmendia L, Hernandez A, Sanchez MB, Martinez JL. Metagenomics and antibiotics. Clinical Microbiology and Infection. 2012 Jul;18.

Leave a Reply