Introduction to Ribosome Profiling
Ribosome-profiling techniques were developed in response to the importance of measuring gene expression at the translational level. Ribosome profiling is a direct technique for determining the precise mRNA region read by the ribosome and for directly examining the transcripts associated with the translation machinery. Ribosome profiling is a cellular snapshot of protein production that has the potential to inform nearly every field of biological and clinical research. In comparison to conventional RNA-Seq studies, ribosome profiling is created to assess mRNA included in inactive translation.
It creates a translatome, which is a "global snapshot" of all the ribosomes actively translating in a cell at a given time. Researchers can now evaluate the location of the translation start sites, the complement of translated ORFs in a cell or tissue, the allocation of ribosomes on messenger RNA, and the rate at which ribosomes translate as an outcome of these findings. Unlike RNA-Seq, which sequences all of the mRNAs of a given sequence in a specimen, ribosome profiling only looks at mRNA sequences that are secured by the ribosome during the translation process. Polysome profiling is not the same as this technique.
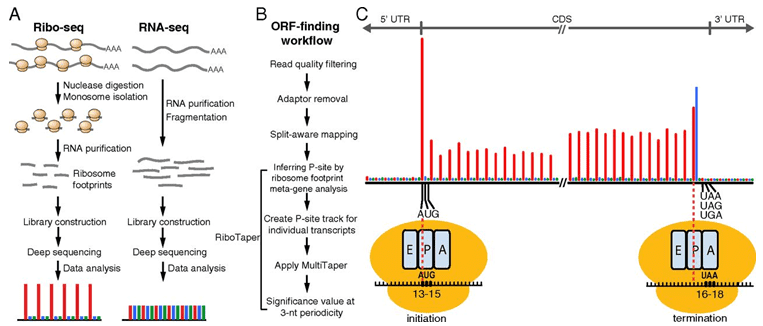
Figure 1. Identifying translated ORFs using ribosome-profiling data. (Hsu, 2016)
Principles of Ribosome Profiling
The reality that translating ribosomes can protect about 30 nucleotides of an mRNA fragment from RNAse digestion allows for ribosome profiling. The traditional molecular method of ribosome footprinting is first used in this technique. In this method, the nuclease is used to destroy unprotected regions of in vitro translated mRNAs. The ribosome-protected mRNAs can be plotted back to the initial mRNA to assess the precise location of the translating ribosome. Ribosome profiling enhances ribosome footprinting by plotting and assessing the whole complement of ribosome footprints to evaluate novel protein synthesis and annotate coding areas globally. mRNA-seq allows for a detailed and accurate analysis of all translating ribosomes thanks to advances in sequencing technology.
Ribosome Profiling Workflow
The translated message can be exactly obtained by evaluating the protected 30 nucleotides during translation using nuclease digestion to isolate nuclease-resistant ribosomes. The procedure for ribosome profiling is as follows:
1. Isolate the mRNA molecules bound to ribosomes by lysing the cells or tissue.
2. Make complexes immobile. This is commonly done with cycloheximide, but it can also be done with other chemicals. With translation-incompetent lysis conditions, it is also possible to avoid using translation inhibitors.
3. Digest the RNA that isn’t protected by ribosomes with ribonucleases.
4. Use sucrose gradient density centrifugation or specialized chromatography columns to isolate the mRNA-ribosome complexes.
5. Purification of the mixture with phenol/chloroform to eliminate proteins.
6. Size-select for mRNA fragments that have been previously protected.
7. Ligate 3′ fragment adapter
8. Subtract rRNA contaminants that have been identified (optional).
9. Reverse transcriptase is used to convert RNA to cDNA.
10. Amplify in a strand-by-strand fashion.
11. Reads from a sequence.
12. determine the translational profile, align the sequence results to the genomic sequence.
Applications of Ribosome Profiling
The three main uses of ribosome profiling are categorizing translated mRNA areas, observing how nascent peptides are folded, and measuring the volume of specific proteins synthesized. Other uses include (1) defining translated sequences within the complex transcriptome, (3) mapping translation initiation areas, (4) measuring differential gene expression at the level of mRNA translation, and (5) identifying novel protein-coding genes and ribosome pausing, and (6) providing data for quantitative RNA sequencing.
References
- Mohammad F, Green R, Buskirk AR. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife. 2019 Feb 6;8.
- Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell. 2016 Mar 24;165(1).
- Hsu PY, Calviello L, Wu HY, et al. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proceedings of the National Academy of Sciences. 2016 Nov 8;113(45).

Leave a Reply