Long Read Sequencing for Epigenomics and Epitranscriptomics
At a glance:
- 1. Introduction
- 2. Overview of Epigenomics and Epitranscriptomics
- 3. Overview of Long-Read Sequencing Technologies
- 4. Application of Long-Read Sequencing in Epigenomics
- 5. Application of Long-Read Sequencing in Epitranscriptomics
- 6. Data Analysis Challenges in Epigenomics and Epitranscriptomics Research
- 7. Future Directions
- 8. Conclusion
1. Introduction
1.1 Importance of Epigenomics and Epitranscriptomics
Epigenomics refers to the study of heritable changes in gene expression and cellular function that do not involve alterations in the DNA sequence itself. Key epigenomic modifications include DNA methylation, histone modifications, and chromatin accessibility. These changes can influence gene activity profoundly, affecting developmental processes, cellular differentiation, and responses to environmental stimuli.
Epitranscriptomics, on the other hand, focuses on RNA-level modifications, such as N6-methyladenosine (m6A), alternative splicing variations, and RNA-binding protein interactions. These modifications play crucial roles in RNA stability, translation efficiency, and cellular adaptation.
1.2 Emergence of Long-Read Sequencing in Epigenetic Research
Traditional sequencing methods, primarily reliant on short-read technologies, face significant limitations in accurately capturing epigenetic modifications and complex transcript structures. Long-read sequencing technologies, particularly those developed by PacBio and Oxford Nanopore, offer direct and comprehensive sequencing capabilities that span entire genomic regions or full-length RNA molecules. This ability to read extended sequences has revolutionized the study of epigenomics and epitranscriptomics, allowing for more detailed insights into regulatory landscapes.
1.3 Core Research Challenges and Application Scenarios
Key challenges in epigenomic and epitranscriptomic research include accurately mapping modifications across the genome, understanding their functional implications, and integrating multi-omics data to elucidate gene regulatory networks. LRS addresses these challenges by enabling high-resolution analysis of methylation patterns, histone modifications, chromatin interactions, and RNA modifications within individual molecules.
1.4 Article Objectives and Structure
This article will outline the distinct roles of epigenomics and epitranscriptomics, detail the technological advancements of long-read sequencing, and explore how these methods are transforming research in these fields. We will compare the strengths and limitations of LRS against traditional methods, providing a comprehensive guide to its application in advanced epigenetic studies.
2. Overview of Epigenomics and Epitranscriptomics
2.1 Definitions and Differences
Epigenomics involves studying DNA-level modifications that affect gene expression without altering the DNA sequence. These include DNA methylation, histone modification, and chromatin accessibility, all of which play crucial roles in gene regulation.
Epitranscriptomics focuses on post-transcriptional RNA modifications that influence RNA processing, stability, and translation. Key modifications include m6A methylation, alternative splicing, and RNA-protein interactions, which are critical for dynamic gene expression regulation.
2.2 Key Components of Epigenomics
- DNA Methylation: Typically occurs at cytosine residues in CpG islands, impacting gene silencing and genomic imprinting.
- Histone Modifications: Covalent modifications such as acetylation, methylation, and phosphorylation alter chromatin structure and gene accessibility.
- Chromatin Accessibility: The organization and compaction of chromatin influence the availability of DNA to transcriptional machinery, directly affecting gene expression.
2.3 Key Components of Epitranscriptomics
- RNA Modifications: m6A is the most prevalent internal modification in eukaryotic mRNA, affecting splicing, export, stability, and translation.
- Splicing Variants: Alternative splicing events lead to diverse protein isoforms, with implications in cellular function and disease states.
- RNA-Binding Proteins (RBPs): RBPs control RNA fate through interactions that influence processing, localization, and degradation.
2.4 Current Research Status and Technical Bottlenecks
Despite advances, current short-read sequencing technologies struggle with accurately mapping epigenetic marks and detecting complex RNA modifications due to read length limitations and amplification biases. Additionally, the resolution needed to capture chromatin conformation and RNA-protein interactions in their native states is often inadequate. Long-read sequencing emerges as a powerful tool to overcome these challenges, providing direct, single-molecule insights into the intricate world of epigenetic and epitranscriptomic regulation.
3. Overview of Long-Read Sequencing Technologies
3.1 Principles of LRS
LRS technologies, such as PacBio's SMRT sequencing and Oxford Nanopore's nanopore sequencing, allow for the reading of long stretches of DNA or RNA in a single pass. These platforms directly detect epigenetic and RNA modifications without requiring PCR amplification or chemical conversion, providing a more accurate representation of the native state of the molecule.
- PacBio SMRT Sequencing: Detects DNA/RNA modifications by monitoring base incorporation kinetics during sequencing.
- Oxford Nanopore Sequencing: Measures changes in ionic current as DNA/RNA passes through a nanopore, detecting base modifications and providing ultra-long reads.
3.2 Comparison with Short-Read Sequencing: Advantages and Challenges
- Advantages:
- Direct Detection of Modifications: LRS can identify methylation and other modifications directly without additional sample preparation steps.
- Comprehensive Coverage: Ability to span repetitive regions and complex structural variants missed by short-read methods.
- Single-Molecule Resolution: Provides insights into heterogeneity and rare events at the single-molecule level.
- Challenges:
- Higher Error Rates: Inherent sequencing errors, though increasingly mitigated by improved algorithms and error-correction techniques.
- Cost and Data Handling: Higher costs and large data sizes necessitate robust computational resources for processing and analysis.
3.3 Unique Value of LRS in Epigenomic and Epitranscriptomic Research
LRS uniquely enables comprehensive mapping of epigenetic modifications and intricate transcript structures, revealing regulatory landscapes that were previously inaccessible. It excels in:
- Whole-Genome Methylation Profiling: High-resolution maps of methylation sites across the genome, crucial for understanding gene regulation in health and disease.
- Chromatin Architecture Analysis: Resolves complex chromatin interactions and topological domains, offering insights into how spatial genome organization influences gene activity.
- Full-Length Transcript Sequencing: Captures complete RNA isoforms, providing detailed views of transcript diversity and regulatory complexity.
Service you may interested in
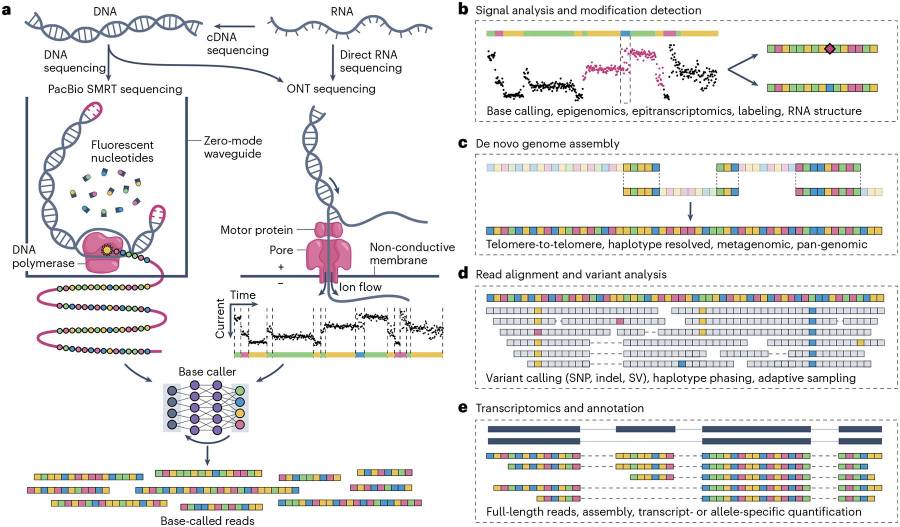 Fig. 1 Long-read sequencing methods and applications.
Fig. 1 Long-read sequencing methods and applications.
4. Application of Long-Read Sequencing in Epigenomics
Epigenomics encompasses the study of heritable changes in gene function that do not involve changes to the DNA sequence itself. DNA methylation, chromatin accessibility, and histone modifications are key components of this regulatory layer, and LRS has significantly advanced the field by providing more comprehensive and accurate mapping of these modifications.
4.1 DNA Methylation Sequencing
Advantages of Long-Read Sequencing in Whole Genome Methylation Detection
DNA methylation, particularly 5-methylcytosine (5mC), plays a critical role in regulating gene expression, genomic imprinting, and X-chromosome inactivation. Long-read sequencing offers distinct advantages over short-read methods in the detection of DNA methylation, including the ability to directly measure methylation without the need for bisulfite conversion. This allows for:
- Full-Length Methylation Profiling: LRS can capture methylation patterns across entire regions of the genome, providing a complete view of epigenetic modifications in repetitive regions and complex loci that are often missed by short-read technologies.
- Single-Molecule Resolution: By sequencing individual DNA molecules, LRS provides a clearer picture of allele-specific methylation and can reveal methylation heterogeneity within a cell population.
- Simultaneous Detection of Multiple Modifications: Unlike traditional methods, LRS can detect multiple types of DNA modifications concurrently, offering a more comprehensive epigenetic profile.
Case Study: Revealing Complex Gene Regulatory Networks
A recent study utilized LRS to map methylation across the entire genome of cancer cells, uncovering previously unrecognized regulatory elements. By analyzing methylation patterns in tandem with gene expression data, the study identified novel regulatory networks that contribute to tumorigenesis. These findings underscore the power of LRS in revealing the intricate relationships between methylation and gene regulation, which are often obscured in studies using short-read sequencing.
4.2 Chromatin Accessibility Analysis
Combining ATAC-seq and Hi-C Technologies with Long-Read Sequencing
Chromatin accessibility plays a pivotal role in gene expression regulation by determining which genomic regions are accessible to transcription factors and other regulatory proteins. Techniques like ATAC-seq and Hi-C have traditionally been used to study chromatin structure and accessibility. When combined with LRS, these technologies can deliver:
- Enhanced Structural Resolution: Long-read sequencing enables the construction of high-resolution chromatin interaction maps, providing insights into the three-dimensional organization of the genome.
- Integration of Epigenetic Data: LRS can simultaneously capture chromatin accessibility and associated DNA modifications, offering a unified view of the epigenetic landscape.
High-Resolution Construction of Chromatin Topological Domains
Long-read sequencing has been employed to build high-resolution maps of chromatin topological domains, such as TADs (topologically associating domains), which play a crucial role in the spatial regulation of gene expression. By capturing long-range interactions and integrating chromatin accessibility data, LRS provides a deeper understanding of the physical organization of the genome and its regulatory impact.
4.3 Histone Modification Analysis
Combining the Advantages of ChIP-seq and Long-Read Sequencing
Histone modifications, such as methylation, acetylation, and phosphorylation, are key regulators of chromatin structure and gene activity. Combining ChIP-seq with long-read sequencing allows for:
- Direct Detection of Histone Marks: LRS can directly detect histone modifications along with associated DNA sequences, enhancing the resolution and accuracy of histone modification mapping.
- Contextual Epigenetic Landscape: This approach provides insights into how histone modifications interact with other epigenetic marks, revealing complex regulatory mechanisms.
5. Application of Long-Read Sequencing in Epitranscriptomics
Epitranscriptomics studies the chemical modifications on RNA molecules, which play critical roles in regulating RNA stability, translation, and splicing. Long-read sequencing offers unique advantages in studying RNA modifications, providing a comprehensive view of the epitranscriptome at single-molecule resolution.
5.1 RNA Modification Analysis
Analysis of Dynamic Changes of RNA Modifications Such as m6A in Different Cell States
RNA modifications, particularly N6-methyladenosine (m6A), are dynamically regulated and influence numerous cellular processes, including differentiation, stress response, and disease progression. Long-read sequencing enables:
- Single-Molecule Detection of Modifications: LRS can directly detect RNA modifications without the need for antibody enrichment or chemical treatment, allowing for unbiased profiling of RNA modifications.
- Temporal Dynamics of Modifications: LRS provides the ability to monitor RNA modifications in real-time and across different cellular states, revealing the dynamic nature of the epitranscriptome.
Research Cases of Single-Molecule Modification Detection Combined with Long-Read Sequencing
A notable application of LRS in epitranscriptomics involved the identification of m6A modifications in stem cells undergoing differentiation. By capturing the full length of modified RNA molecules, researchers were able to correlate specific modifications with changes in gene expression, highlighting the role of m6A in regulating cell fate decisions.
5.2 Spliceosome Analysis
Advantages of Long-Read Sequencing in Identifying Complex Splicing Variants
Alternative splicing significantly contributes to proteome diversity, and aberrant splicing is implicated in various diseases. Long-read sequencing provides distinct benefits in spliceosome analysis, including:
- Identification of Full-Length Isoforms: LRS captures entire RNA molecules, allowing for the accurate detection of complex splicing events that are often missed by short-read sequencing.
- Direct Detection of Splice Junctions: This technology enables the identification of novel splice junctions and rare splicing events, providing a more comprehensive view of the transcriptome.
Dynamic Monitoring of Mature RNA and Precursor RNA
LRS has been instrumental in monitoring the dynamics of splicing, including the transition from precursor to mature RNA. This real-time observation provides insights into the kinetics of splicing and how splicing errors contribute to disease.
5.3 RBP Interaction
Innovative Application of Long-Read Sequencing in Analyzing RNA-RBP Interaction
RNA-binding proteins play crucial roles in post-transcriptional regulation by interacting with RNA molecules. Long-read sequencing allows researchers to:
- Map RNA-Protein Interactions at Single-Molecule Resolution: LRS can directly capture RNA-RBP interactions without cross-linking, providing a more accurate representation of in vivo interactions.
- Integration of Epitranscriptomic Data: Combining RNA modification data with RBP interaction profiles offers a holistic view of post-transcriptional regulation.
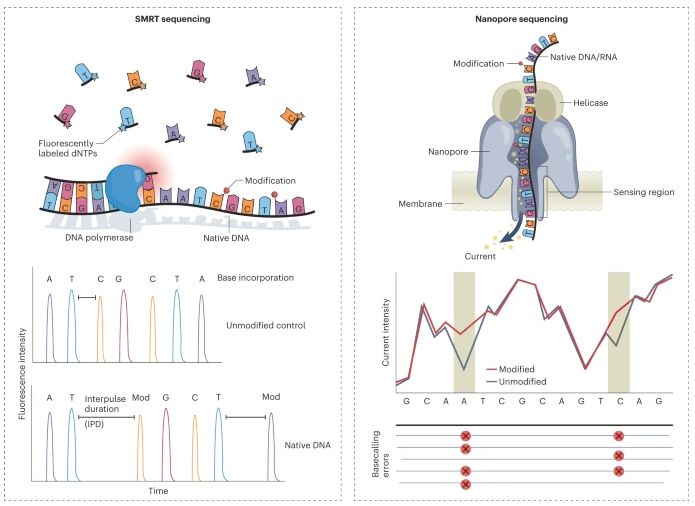 Fig. 2: Overview of methods for detecting DNA and RNA modifications using long-read sequencing.
Fig. 2: Overview of methods for detecting DNA and RNA modifications using long-read sequencing.
6. Data Analysis Challenges in Epigenomics and Epitranscriptomics Research
Despite the transformative potential of LRS, significant challenges remain in the analysis of the vast and complex data it generates.
6.1 Big Data Processing: Correction and Accurate Alignment of Long-Read Sequencing Data
- Error Correction: LRS technologies, while powerful, have higher error rates compared to short-read sequencing. Advanced algorithms and hybrid approaches that integrate short reads are often required for error correction.
- Accurate Alignment: Aligning long reads to the reference genome is computationally intensive, especially in repetitive or highly modified regions. Improved alignment tools are essential for accurate data interpretation.
6.2 Data Integration and Multi-Omics Analysis
- Integrative Analysis: Combining LRS data with other omics datasets (e.g., proteomics, metabolomics) provides a comprehensive view of cellular states but requires sophisticated bioinformatics tools capable of multi-dimensional data integration.
- Cross-Platform Comparability: Variability between sequencing platforms can complicate data integration. Standardized protocols and cross-validation methods are needed to ensure consistent and reproducible results.
6.3 Bioinformatics Tools: Current Software and Performance Comparisons
- Tool Selection: A wide array of bioinformatics tools exists for LRS data analysis, each with varying performance. Tools such as Nanopolish, Guppy, and Nextflow offer distinct advantages depending on the specific application, but their performance in terms of accuracy, speed, and scalability needs careful consideration.
- Scalability and Usability: As LRS datasets grow larger, the need for scalable, user-friendly bioinformatics solutions becomes more pressing. Advances in machine learning and cloud computing offer potential pathways to address these challenges.
7. Future Directions
7.1 Emerging Technological Trends: Single-Cell Epigenomics, Time-Series Sequencing, and Beyond
Single-Cell Epigenomics: LRS is rapidly expanding into single-cell applications, allowing researchers to probe epigenetic heterogeneity at the individual cell level. Techniques such as single-cell DNA methylation sequencing and single-cell ATAC-seq combined with LRS provide high-resolution maps of epigenetic states across diverse cell populations, revealing new insights into cellular differentiation, disease progression, and tissue heterogeneity.
Time-Series Sequencing: Time-series epigenomic and epitranscriptomic studies using LRS enable the tracking of dynamic regulatory changes over time. This approach is particularly valuable for understanding transient epigenetic modifications and RNA processing events that occur during development, disease, and environmental adaptation.
Integration with Novel Sequencing Modalities: Emerging methods such as co-sequencing of DNA and RNA from the same molecule (e.g., DR-seq) using LRS platforms are pushing the boundaries of integrated multi-omics analysis, offering a deeper understanding of the interplay between genetic, epigenetic, and transcriptomic regulation.
7.2 Clinical Applications: Constructing Disease Epigenomic Maps and Personalized Therapeutics
Disease Epigenomic Mapping: LRS is instrumental in constructing comprehensive epigenomic maps of various diseases, including cancer, neurological disorders, and autoimmune conditions. By capturing complex epigenetic alterations, LRS enables the identification of novel biomarkers and therapeutic targets.
Personalized Medicine: The high resolution and single-molecule precision of LRS support the development of individualized epigenetic profiles, paving the way for personalized treatment strategies. Epigenomic and epitranscriptomic data obtained from LRS can guide the selection of targeted therapies, monitor treatment responses, and predict disease relapse.
7.3 Cross-Disciplinary Integration: Applications with Machine Learning and Artificial Intelligence
Machine Learning in Epigenomic Data Analysis: The integration of LRS data with machine learning algorithms enhances the ability to predict gene regulatory networks, identify epigenetic signatures, and classify disease states. AI-driven models can analyze complex datasets to uncover hidden patterns that are not apparent through conventional analysis methods.
AI-Enhanced Error Correction and Variant Calling: AI tools are being developed to address the high error rates in LRS by improving the accuracy of base calling, variant detection, and methylation status identification. These advancements are critical for translating LRS data into actionable biological insights.
Robust Data Integration: AI-driven data integration methods facilitate the combination of LRS with other omics datasets, helping to decode multifaceted biological processes. This approach supports the discovery of synergistic interactions between epigenetic modifications, gene expression, and protein function.
8. Conclusion
Key Findings and Highlights
Revolutionizing Epigenomics and Epitranscriptomics: Long-read sequencing technologies are redefining our understanding of the epigenome and epitranscriptome by providing unprecedented resolution, direct detection capabilities, and the ability to analyze complex regulatory networks in full detail.
Applications Across Multiple Domains: From whole genome methylation sequencing and chromatin accessibility analysis to RNA modification and spliceosome dynamics, LRS enables comprehensive profiling of the regulatory landscapes that drive cellular function and disease.
Overcoming Data Challenges: Although significant challenges remain in data processing, error correction, and integration, ongoing advancements in bioinformatics tools and AI-driven approaches are poised to address these hurdles, further enhancing the utility of LRS.
The Role of Long-Read Sequencing in Driving Innovation
LRS is a powerful tool that is propelling the fields of epigenomics and epitranscriptomics into new realms of discovery. By capturing the intricacies of epigenetic and RNA modifications at an unparalleled scale, LRS allows researchers to explore complex biological questions with greater depth and precision than ever before.
Encouraging Continued Exploration and Innovation
The future of LRS in epigenomics and epitranscriptomics is bright, with ongoing research poised to unlock even deeper insights into the regulatory mechanisms that underlie health and disease. Continued innovation, interdisciplinary collaboration, and the development of new technologies will be essential in fully realizing the potential of LRS, shaping the future of precision medicine, and transforming our approach to understanding the molecular underpinnings of life.
References
- Lucas, M.C., Novoa, E.M. Long-read sequencing in the era of epigenomics and epitranscriptomics. Nat Methods 20, 25–29 (2023).
- Kovaka S, Ou S, Jenike KM, Schatz MC. Approaching complete genomes, transcriptomes and epi-omes with accurate long-read sequencing. Nat Methods. 2023 Jan;20(1):12-16.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment