The human gastrointestinal tract is colonized by a vast array of microbes, which form communities of bacteria, viruses and microbial eukaryotes that are specific to people of different ages, diet habits, and health conditions. Many of these microbes have neither been cultured independently nor studied thoroughly. Whole-genome de novo sequencing refers to sequencing a novel genome with no requirement of reference sequences, which allows for culture-independent sophisticated analysis and sampling of gut microorganisms. The symbiotic relationships between gut microorganisms and the host can be mutualism, commensalism or parasitism. Generally, gut microbiota maintains dynamic balancing when our bodies function well. While dysbiosis might occur due to internal or external causes, where the influence could be bilateral vice versa. In addition, the appearance or over-proliferation of parasitic microbes could also be the unignorable inducement for the breakdown of community stability and the reason for jeopardizing our health, making it pivotal to study certain disease-related microbes and their relationship between human.
Unlike approaches that only targeted a limited portion of the genome such as 16S/18S/ITS amplicon sequencing, whole-genome sequencing delivers a comprehensive view of the entire genome, which works as a powerful tool in multiple areas including diagnosis, tracking disease outbreaks, characterizing causative mutations and identifying drug-resistance genes.
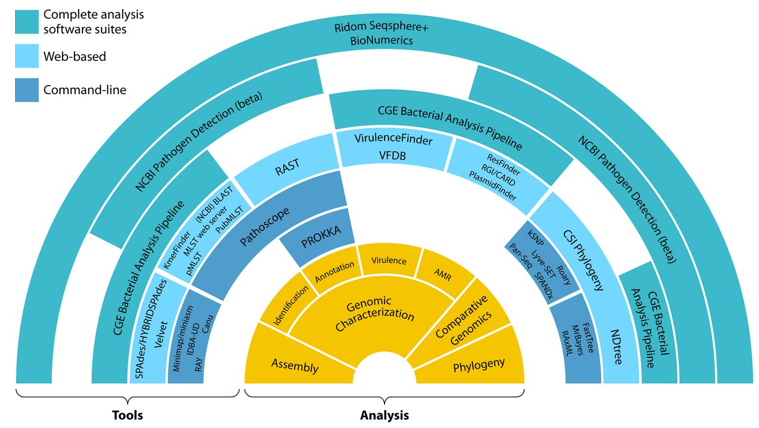
Figure 1. Whole-genome sequencing analysis tools (Quainoo 2017)
CD Genomics provides advanced whole-genome sequencing services, the workflow generally falls into the following steps:
The feature processes of whole-genome sequencing workflow are alignment, variant calling, annotation and calculating related metrics. In the alignment process, we have access to up-to-date reference databases to guarantee a detailed, accurate comparison. We are capable of running multiple genetic variant calling, such as single nucleotide variants (SNVs), indels calling, copy number variants (CNVs), and structural variants (SVs), which is the process to identify genetic variants that can subsequently be associated with important phenotypes. Then followed by data analysis and variant annotation, including closed-reference operational taxonomic unit (OTU) analysis, genus-level taxonomy analysis, diversity analysis, species-level taxonomy analysis, KEGG metabolic pathway and module analysis, etc., to fully understand the genome sequence. Finally, is the calculation of alignment and related metrics.
Sample Requirements
| Sample Type | Concentration | A260/280 | Minimum Amount |
| Genomic DNA | 50 ng/µl minimum | 1.8-2.0 | 1 µg |
Our Advantages
◆ High coverage and highly sensitive
◆ Wide read range
◆ Cost-efficient and fast turnaround time
◆ Bioinformatics support
◆ Specialized in gut microbiota analyses
We provide a full package of genomics sequencing services not just limited to the processes above, other optional informatics services and custom analysis are also available. If you are interested in our whole-genome sequencing services and want to find out more, feel free to contact us!
Reference
1 Quainoo S; et al. Whole-Genome Sequencing of Bacterial Pathogens: the Future of Nosocomial Outbreak Analysis. Clinical Microbiology Reviews. 2017, 30(4): 1015.
*For Research Use Only. Not for use in diagnostic procedures.