The viruses that make up the intestinal virome are almost equal to the number of bacterial cells. Each gram of the human gut contains approximately 108–109 virus-like particles (VLPs), the majority of which are cross-assembly phages belonging to the family Podoviridae. Viruses interact with other organisms within the microbiome where they can eventually change the host phenotype causing either beneficial or harmful effects to the organism because of their ability to directly influence disease susceptibility and resistance. Currently, only 1% of the virome has been sequenced, leaving the majority of the virome yet to be identified.
Virus Sequencing
Many methods can be applied for virus identification. Older references relied on electron microscopy and virulence specificity methods. Modified extraction and initial random amplification made microarray sensitive for all virus identification studies. Currently, sequencing is an accurate, efficient, and the most widely used method for virus identification. Hybridization and PCR-based methods are effective for enrichment, which largely reduce nucleic acid contamination and sequencing depth.
About Our Service
In CD Genomics, our NGS-based gut virus identification aims to identify species efficiently in a fast and affordable high-throughput, and simplified manner. We prioritize delivering high-accuracy and high-sensitivity data. Our services have advantages such as aseptic operation during the experiment, user-friendliness, accuracy and timeliness, high-throughput, detection of novel viral strains, and meet the requirements of identification and research.
Taxonomic assignment of microbial species in complex network analysis, microbial communities, evolutionary studies, and correlation analysis is made possible through NGS-based microbial identification either by single genetic targets or whole-genome strategies.
Our Advantages:
- Hybridization/amplification-based enrichment methods allow for cost-effective identification
- Next-generation sequencing provides a culture-free process that helps the entire microbial population to be analyzed within a specimen
- NGS gives the capacity to integrate multiple specimens
- High sensitivity and accuracy
- Affordable price and professional team
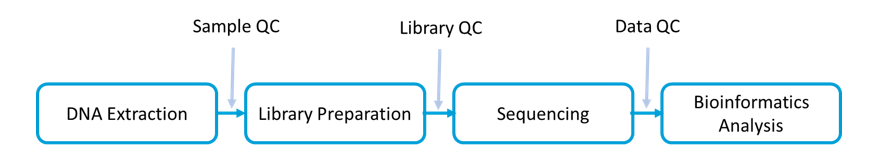
Workflow and Bioinformatics Analysis
| Bioinformatics Analysis | Details | |
| Routine identification analysis | Morphological, physical, and chemical properties of the sample | |
| Composition analysis | Rank-abundance curve | Detects abundance and distribution of the species in the sample |
| OUT classification | Species and genera identification | |
| Diversity analysis | Alpha diversity | Diversity of species in an ecosystem |
| Beta-analysis | Comparative studies between samples | |
| Comparative analysis | PCA | Correlation between microbial populations |
| NMDS analysis | Similarity and dynamism of microbial sample through time | |
| LDA | Clustering between study groups | |
| Evolutionary analysis | Phylogenetic tree construction | |
CD Genomics provides gut virus profiling services at high depth with high precision to meet the needs of studying population genetics and microbial profiles in the gastrointestinal tract, in order to research into biodiversity within and interaction between these viruses and human bodies. Our enrichment-based virus profiling protocol uses hybridization and amplification to study the viruses of your interest. If you are interested in our amplicon sequencing service and want to find out more, feel free to contact us!
Reference:
1. Bexfield N, Kellam P. Metagenomics and the molecular identification of novel viruses. The Veterinary Journal. 2011, 190(2).
*For Research Use Only. Not for use in diagnostic procedures.