The totality of fungi, viruses, bacteria, and protozoa, and their entire genetic material contained in the gastrointestinal tract (GIT) describes the gut microbiome of an organism. It is imperative to support studies defining microbial diversity in varying human populations. Results could help design future therapeutic methods for the restoration of the meta- Human organism wellness.
The gut microbial diversity analysis is usually anchored on the 16S/18S/ITS amplicon sequencing platform. It enables efficient characterization of microbial diversity by utilizing next-generation sequencing technology and other molecular techniques. CD Genomics experts utilize this platform to greatly assist our customers and meet regulatory requirements at the same time. Our team consists of around 70% of scientists with a Ph.D., and our advanced sequencing platforms such as our PacBio systems and Illumina PE250/300 are available to cater to varying sequencing requirements.
About Our Service
Our innovative microbial diversity detection platform produces an accurate and high-throughput analysis of the 16S/18S/ITS region via combined advanced sequencing technology and quantitative PCR (qPCR). Both short-read 16S/18S/ITS sequencing by next-generation sequencing (NGS) technology and full-length 16S/18S/ITS sequencing by PacBio SMRT sequencing and Nanopore sequencing can be delivered. Sanger sequencing or qPCR with genus- and species-specific primers are used for validating the results and to quantify microbial species. Prokaryotic diversity analysis and eukaryotic diversity analysis can be both conducted in our gut microbial diversity platform utilizing the role of 16S/18S/ITS as the molecular marker in prokaryotes and eukaryotes.
Our Advantages:
- Detection of low abundance species of microbial communities
- Years of commercial experience in life sciences
- Quick and affordable services
- Cutting-edge sequencing platforms featuring both next-generation and long-read sequencing platforms
- Flexible sequencing strategies and data analysis.
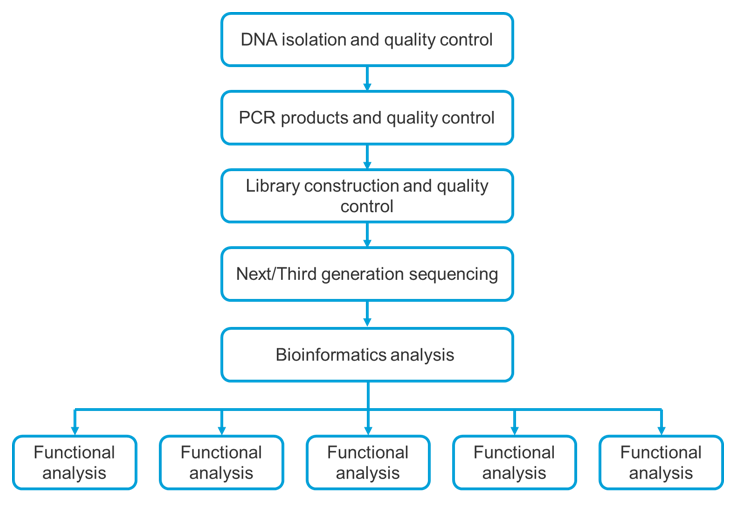
Gut Microbial Diversity Analysis Workflow
Bioinformatics Analysis
CD Genomics provides customizable one-stop service to deliver quality analysis and meet your preferences. The bioinformatics analysis which is divided into five parts: Data QC, genome assembly, prediction of functional elements, gene function annotation, and comparative genome analysis.
| Bioinformatics Analysis | Issues Addressed | |
| Species annotation | Species annotation and OUT clustering | |
| Diversity Analysis | α diversity | Species diversity in one microbial ecosystem |
| β diversity | Microbial community diversity between different environments | |
| Meta-analysis | Comparative studies from large datasets from different sources | |
| Evolutionary analysis | Establishment of a phylogenetic tree | |
| Functional analysis | intestinal microflora analysis | Loci detection of gut bacteria, disease prediction and treatment |
| PICRUSt | Functional profile prediction | |
| FAPROTAX | Biochemical cycle prediction | |
| Tax4fun | Functional profile prediction | |
| FunGuild | Ecological role prediction | |
| Correlation analysis | CCA | Correlation between microbes and environment |
| VPA | ||
| Network analysis | ||
Sample Requirements
Before sending us your samples, make sure they meet the following requirements: OD260/230 ≥ 1.8, 1.8 < OD260/280 < 2.0 without degradation or contamination. In Illumina, amount of DNA must be ≥ 300 ng with PCR Products at ≥ 400 ng. In PacBio, gDNA must be at ≥ 100 ng with PCR Products yielding ≥ 400 ng.
CD Genomics provides gut microbial diversity analysis service at high accuracy in hoping to lead to more valuable insights, in order to research into biodiversity within and interaction between these microbes and human bodies. Our platforms include Illumina, Pacbio SMRT, and Oxford Nanopore to meet your needs no matter what and how long your interested targets are. If you are interested in our amplicon sequencing service and want to find out more, feel free to contact us!
References:
1. Cresci GA, Izzo K. Gut Microbiome. InAdult Short Bowel Syndrome. Academic Press, 2019.
2. Das B, Ghosh TS, Kedia S, et al. Travis SP. Analysis of the gut microbiome of rural and urban healthy Indians living in sea level and high altitude areas. Scientific reports. 2018, 8(1).
*For Research Use Only. Not for use in diagnostic procedures.