The human gastrointestinal microbiota represents one of the most complex ecosystems on earth. We now have the consensus that bacterial microbiota communities have been established since our birth and evolve over time to become 'adult-like' bacterial communities. DNA and RNA viruses, which include eukaryotic viruses, endogenous retroviruses, bacterial viruses and archaeal viruses, collectively make up the intestinal virome outnumber bacterial cells by as much as 10:1. Despite its predominance, our knowledge of gut viral communities is relatively deficient, as to both the acquisition and evolution.
Considering that viruses only replicate inside living cells, it is obvious that virome should not be studied in isolation. The human gut virome includes a diverse collection of viruses that infect our own cells as well as other commensal organisms, plays an important role in homeostatic regulation and disease progression, and directly or indirectly impact our well-being.
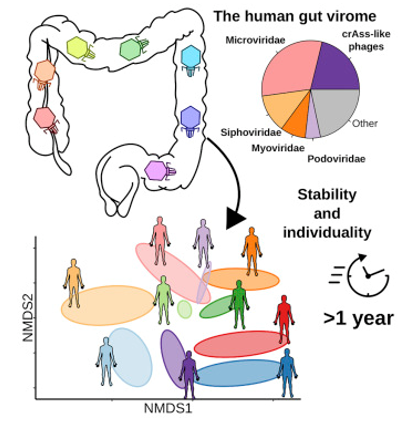
Figure 1. The diversity and stability of gut virome in humans. (Shkoporov 2019)
Due to the lack of universal marker, identifying virome information in large datasets is much more challenging than the cellular counterparts, which can easily be obtained through homology sequence searches against reference databases. In contrast to the sequencing of cellular organisms, extra sample preparation procedures are needed to enrich and purify viral particles, including filtration, ultracentrifugation, nuclease treatment, multiple displacement amplification (MDA), linker-amplified shotgun library (LASL), and random PCR approach. Along with the advances in high-throughput and deep sequencing technology, we are capable of characterizing virome richness and stability, gene functions, and association with disease phenotypes, which also helps to understand the richness of the virome and the role viruses play within complex microbial communities.
Sample Requirements
| Sample Type | Concentration | Minimum Amount |
| DNA / cDNA | 50 ng/µl minimum | 3 µg |
Data Analysis
◆ Operational taxonomic unit (OTU) binning
◆ Taxonomy assignment
◆ Abundance table construction
◆ Compositional data analysis (CoDA)
◆ Diversity analysis
◆ Principal coordinates analysis (PCoA)
◆ Multivariate differential abundance testing
◆ Data annotation
◆ Other analyses
Our Advantages
◆ All types of viruses (ssDNA viruses, dsDNA viruses, ssRNA viruses, ds RNA viruses, retroviruses, etc.)
◆ Highly sensitive and accurate
◆ Wide read range
◆ Cost-efficient and fast turnaround time
◆ Bioinformatics support
◆ Specialized in gut microbiota analyses
CD Genomics is one of the most trusted companies in the field, we strive to provide the best strategies for your gut virome studies. We provide full support for from fine sample preparation to advanced sequencing, then to deep data analysis, and our customized service will guarantee the high quality of your results and provide more sparkling ideas for your research. To find more about virome sequencing for your gut microbiota research, please feel free to contact us.
Reference
1. Shkoporov AN; et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host & Microbe. 2019, 26(4).
*For Research Use Only. Not for use in diagnostic procedures.