A wide range of microbes, which include bacteria, viruses, and microbial eukaryotes that are particular to different age groups, diet habits, and health issues, colonize the human gastrointestinal tract. The gut microbiota is critical to human health and fitness. Regardless of the fact that metagenomic research offered us a mass of useful information, bacterial species cannot be thoroughly analyzed without analyzing protein expression under specific environmental factors. Proteogenomics is a branch of biology that employs a variety of proteomics, genomics, and transcriptomics to assist in peptide exploration and classification. Proteogenomics is a technique for discovering new peptides by evaluating MS/MS spectra to a protein database obtained from genomic and transcriptomic information. Proteogenomics often involves a study that utilizes proteomic results to better gene annotations, often obtained from mass spectrometry. Genomics is the study of entire organisms' genetic code, whereas transcriptomics is the study of RNA sequencing and transcripts.
About Our Proteogenomics Service
As a prominent company with many years of proteomics knowledge, CD Genomics offers state and most feasible mass spectrometry technologies to add significance to your study on intestinal metaproteomics. By obtaining proteomic information, our proteogenomics service can greatly improve gene annotations. The first of the two strategic paths we can offer is two-dimensional electrophoresis associated with mass spectrometry; the second is chromatography integrated with mass spectrometry, also referred to as the shotgun technique. Both strategies involve the utilization of mass spectrometry, which performs detection as well as composition and structural analysis by determining the mass to charge ratio (m/z) of proteins.
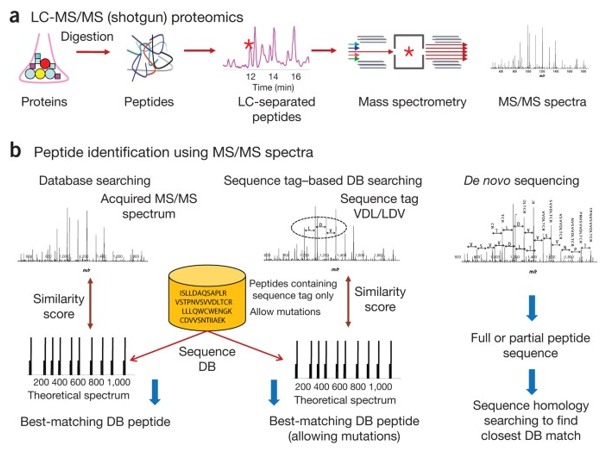
Figure 1. Proteogenomics Analysis by LC-MS/MS and MS/MS. (Nesvizhskii, 2014)
Our Advantages
- Exceptional sensitivity
- Enormous sequence, structure, and function information
- Availability to both large biological and small organic molecules
- Peptides and protein sequence assessment
- Obtain enormous quantities of information
Workflow and Bioinformatics Analysis
| Analysis Content | Details |
| Functional annotations | Assessing individual proteins in datasets and allocating each one a specific function |
| Protein database searching and annotation | To conduct referencing data assessment in both public and commercial datasets to ensure the completeness and accuracy of the comparison and annotation. Also accessible are de novo searching and homology searching. |
| Quantitative taxonomic analysis | For comparative metaproteomics research, protein quantification is essential. Quantification will mainly show the level of the microbiota from a taxonomic point of view. |
| Clustering | On the grounds of a statistical distance configuration, dividing observed databases into a few subclasses or clusters. |
| Metabolic pathways linking analysis | The assessment of particular proteins to selected metabolic processes will help researchers better comprehend main cellular signaling and metabolic networks, as well as reveal microbiota biological frameworks. |
References
1. Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nature methods. 2014, 11(11).
2. Aires J, Butel MJ. Proteomics, human gut microbiota and probiotics. Expert review of proteomics. 2011, 8(2).
3. Denef VJ, Kalnejais LH, Mueller RS, et al. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proceedings of the National Academy of Sciences. 2010, 107(6).
4. Delmotte N, Knief C, Chaffron S, et al. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proceedings of the National Academy of Sciences. 2009, 106(38)
*For Research Use Only. Not for use in diagnostic procedures.