Cas-Offinder: An Online Tool for sgRNA Off-Target Prediction
With its straightforward design, efficiency, and low cost, the CRISPR/Cas9 system has emerged as a pivotal tool in the field of gene editing. This system consists of two core components: the Cas9 nuclease and the guide RNA (sgRNA). The sgRNA plays a crucial role by accurately recognizing specific Protospacer Adjacent Motifs (PAM) adjacent to the target DNA sequence. By closely binding with the Cas9 protein, sgRNA directs Cas9 to perform precise cuts in the target DNA. However, if the sgRNA inadvertently binds to non-target sequences in the genome, off-target effects may arise, potentially compromising experimental outcomes.
Consequently, designing sgRNAs that are both efficient and highly specific, while effectively predicting their potential off-target effects, has become a critical issue within the scientific community. To address this need, we recommend an online tool—Cas-Offinder (http://www.rgenome.net/cas-offinder/). This tool is specifically designed for sgRNA off-target prediction and includes comprehensive operating guidelines, aiming to facilitate researchers in conducting relevant studies with greater ease and accuracy.
Overview of Cas-Offinder
Cas-Offinder is a sgRNA off-target prediction tool developed by the Kim team at Seoul National University in South Korea, noted for its unique functionality. Its operational workflow can be summarized in several steps:
- Users are required to input key information about the on-target sgRNA, including the PAM sequence, length, and species information.
- Cas-Offinder then automatically scans the entire genome to identify candidate sequences that share the same PAM sequence and length as the on-target sgRNA.
- The tool conducts a detailed comparison between the identified sequences and the on-target sgRNA, calculating key metrics such as mismatches, insertions, and deletions.
- Finally, based on user-defined thresholds (including acceptable ranges for mismatches, insertions, and deletions), Cas-Offinder intelligently filters the results to identify potential off-target sequences, which are subsequently output for further analysis.
This tool provides researchers with a reliable method for accurate sgRNA design and off-target effect prediction, thereby supporting the robust development of gene editing technologies.
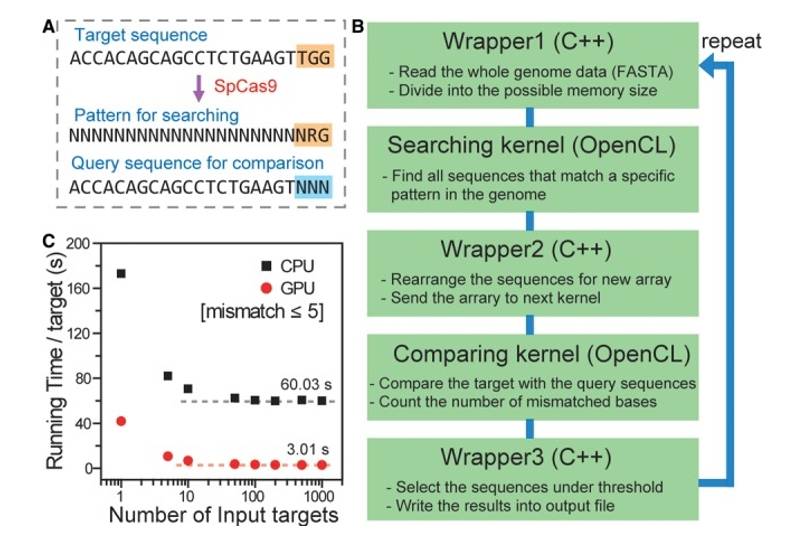 Fig. 1. (A) The scheme of Cas-OFFinder. (B) The workflow of Cas-OFFinder. (C) Running time per target site as a function of the number of input target sites via CPU (black squares) and GPU (red circles)
Fig. 1. (A) The scheme of Cas-OFFinder. (B) The workflow of Cas-OFFinder. (C) Running time per target site as a function of the number of input target sites via CPU (black squares) and GPU (red circles)
You may interested in
CD Genomics offers a range of services related to gene editing predictions and off-target detection, including:
Take the Next Step: Explore Related Services
Cas-Offinder Operation
Input Requirements for Cas-Offinder
To utilize Cas-Offinder effectively, users must provide the following six key pieces of information:
- Type of PAM sequence;
- Species;
- sgRNA sequence to be predicted;
- Maximum allowable number of mismatches for off-target sequences;
- Maximum allowable length of insertions (DNA bulges) for off-target sequences;
- Maximum allowable length of deletions (RNA bulges) for off-target sequences.
The platform includes pre-set options for common PAM sequence types and species to facilitate user selection. For specific requirements, users are encouraged to contact the tool's developers to request the addition of new sequence types or species. Alternatively, users may download the Cas-Offinder code and execute it via command line to accommodate personalized needs.
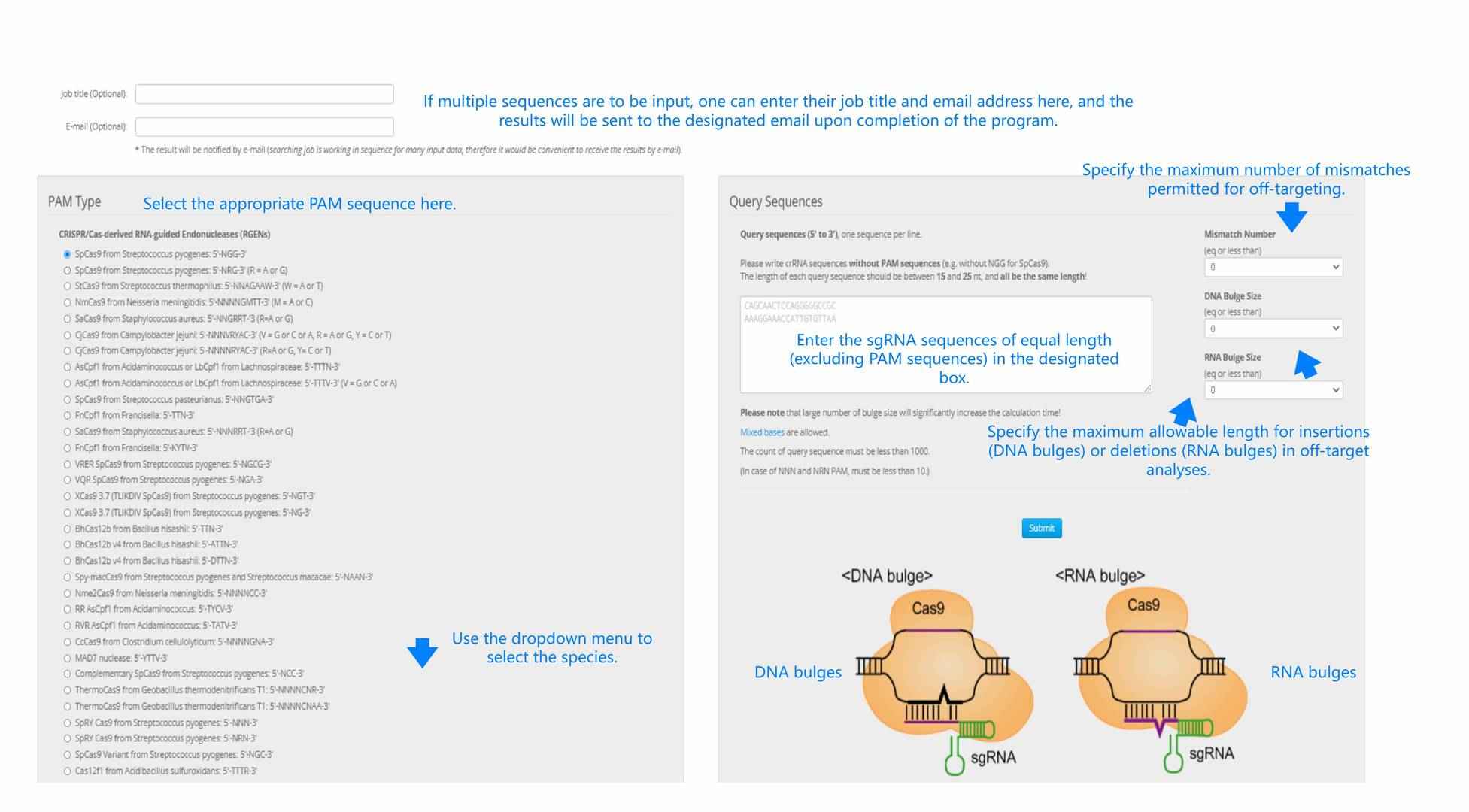 Figure 2. An illustration of the input requirements for Cas-Offinder.
Figure 2. An illustration of the input requirements for Cas-Offinder.
If multiple sequences are entered, users can provide their job title and email address here; the program will send the results to the specified email upon completion.
Select the appropriate PAM sequence from the options provided.
Choose the species from the dropdown menu.
Specify the maximum allowable number of mismatches for off-target sequences.
Enter the sgRNA sequences of equal length (excluding the PAM sequence) in the designated box.
Indicate the maximum allowable lengths for insertions (DNA bulges) or deletions (RNA bulges).
Cas-Offinder Output
Cas-Offinder presents the predicted results primarily in two tables: a summary table and a detailed table.
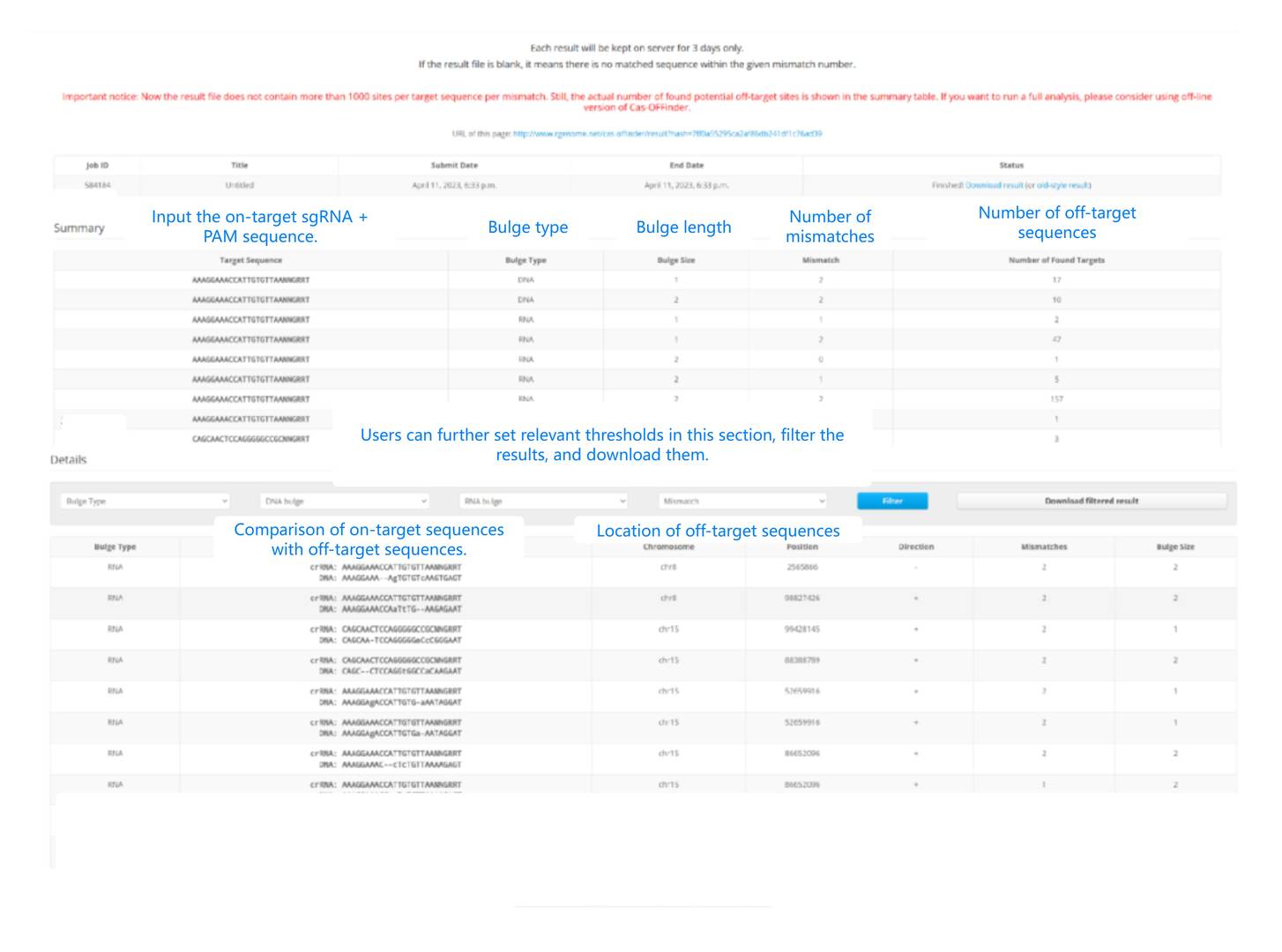 Figure 3. An explanation of the Cas-Offinder output results.
Figure 3. An explanation of the Cas-Offinder output results.
Cas-Offinder demonstrates significant advantages in predicting off-target effects, as its prediction process is not constrained by PAM sequences. This flexibility allows it to effectively address various mutations within DNA sequences, including mismatches, insertions, and deletions, thereby showcasing its robustness and practicality. However, it is important to note that while Cas-Offinder can identify potential off-target sites, the actual probability of off-target events occurring at these locations remains uncertain.
References
- Bae S., Park J., & Kim J.-S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473-1475 (2014).

