Bisulfite conversion is a process of treating DNA with bisulfite, which can transform unmethylated cytosines to uracils while leaving methylated cytosines unaltered. The unaltered Methylated cytosines can be calculated after sequencing and bioinformatics analysis. The CD DNA Methylation Bisulfite Kit was created specifically for bisulfite conversion of DNA in a quick and easy manner. Combining DNA denaturation (by thermal denaturation rather than chemical denaturation) and bisulfite conversion into a single step simplifies the operation procedure and cuts experiment time in half. The unmethylated cytosine conversion efficacy is no less than 99 percent, and the recovery effectiveness is no less than 80 percent. Endonuclease digestion, sequencing, microarrays, and other downstream analyses can utilize the purified and converted DNA immediately.
Storage:
Store at room temperature (15-25℃) . The dissolved CT Conversion Mix can be stored at room temperature (15°C ~ 25°C) for 24 h, frozen (-30°C ~ -15 °C) for 1 month; and protect it away from light.
Components:
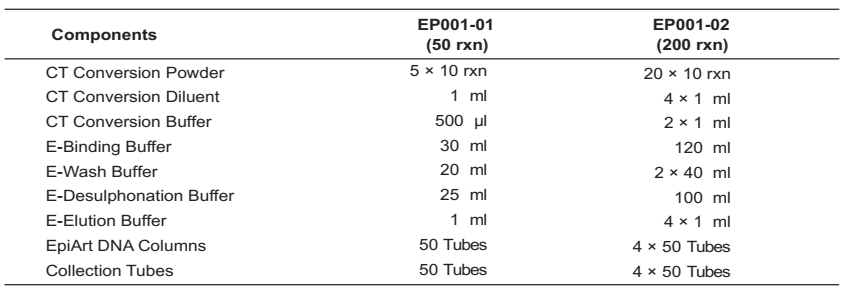
Specifications:
| Sample amount | 100pg - 2μg |
| Features | Combine DNA denaturation (by thermal denaturation instead of chemical denaturation) and bisulfite conversion into one step, simplified operation process and reduced experiment time. The recovery efficiency no less than 80% and the unmethylated cytosine conversion efficiency is no less than 99%. |
| Application | The purified and converted DNA can be used directly for downstream analyses including endonuclease digestion, sequencing, microarrays, etc. |
| Species Category | animals, plants, microorganisms |
| Sample type | genomic DNA, cell-free DNA (cfDNA) and Amplicons from cells or tissues |
| Sequencing Platform | Illumina |
Reagent Preparation
- Please add proper volume of absolute ethanol to E-Wash Buffer before first use (Add 80 ml of absolute ethanol to E-Wash Buffer in EP001-01; add 160 ml of absolute ethanol to each bottle of E-Wash Buffer in EP001-02). After adding ethanol, tighten the lid tightly to prevent evaporation of ethanol.
- Prepare CT Conversion Mix:
Add 1 ml of ddH2O, 200 μl of CT Conversion Diluent and 100 μl of CT Conversion Buffer to a tube of CT Conversion Powder. Vortex at room temperature for about 1 min until dissolved. Each tube of CT Conversion Mix can be subjected to about 10 reactions.
Bisulfite Conversion
- Equilibrate CT Conversion Mix to room temperature and prepare the reaction solution in a 200 μl sterile PCR tube.
- Mix the tube by inverting or pipetting, then centrifuge briefly to collect the reaction solution to the bottom of the tube.
- Put the tube in a PCR instrument and run the PCR program.
Purification of Conversion Products
- Assemble the EpiArt DNA Columns with the Collection Tubes.
- Add 600 μl E-Binding Buffer to the EpiArt DNA Column, then add the reaction solution from Step 08-2/ to the adsorption column; gently mix the tube by inverting for 8-10 times to mix thoroughly the reaction solution with E-Binding Buffer.
- Centrifuge at 12,000 × g for 30 - 60 sec, discard the filtrate and reuse the Collection Tube.
- Add 500 μl of E-Wash Buffer (make sure that the absolute ethanol has been added before use) to the adsorption column. Centrifuge at 12,000 × g for 30 - 60 sec, discard the filtrate and reuse the Collection Tube.
- Add 500 μl E-Desulphonation Buffer to the adsorption column, incubate at room temperature (15°C - 25°C) for 15 min, centrifuge at 12,000 × g for 30 - 60 sec, then discard the filtrate and reuse the Collection Tube.
- Add 500 μl of E-Wash Buffer (make sure that the absolute ethanol has been added before use) to the adsorption column. Centrifuge at 12,000 × g for 60 sec, discard the filtrate and reuse the Collection Tube.
- Repeat Step 6.
- Centrifuge empty adsorption column at 12,000 × g for 2 min.
- Transfer the adsorption column to a new 1.5 ml centrifuge tube, open the lid for 2 minutes and dry thoroughly. Add 10 µl -20 µl of E-Elution Buffer to the center of the adsorption column, then place it at room temperature for 1 - 2 min, centrifuge at 12,000× g for 2 min to collect DNA filtrate.
- Discard the adsorption column. Store the DNA at -20°C and stored at -70 °C for long-term.



