The CD rRNA Depletion Kit (Bacteria) is a kit for eliminating ribosomal RNA (16 S and 23 S rRNA) from Gram-negative and Gram-positive bacteria. This kit allows you to remove rRNA from 1g to 5g of undamaged or deteriorated total RNA. The RNA collected can be utilized to generate a transcriptome library with strand-specific information. The kit includes rRNA hybridization probe, RNase H digestion, DNaseI digestion, buffers, and other reagents required for rRNA depletion. Regarding the digestion stages, the retained mRNA and other non-coding RNA can be utilized to create an RNA library that can be used to analyze LncRNA and other non-coding RNAs.
Storage:
All the components should be stored at -20℃.
Components:
-Components.png)
Specifications:
| Sample amount | 1µg -5µg |
| Features | Compatible with a wide range of Gram-negative and Gram-positive bacteria. Remove 16 S and 23 S rRNA effectively. |
| Application | The obtained RNA can be used for transcriptome library construction with strand-specific information. |
| Species Category | compatible with Gram-negative and Gram-positive bacteria |
| Sample type | intact or degraded total RNA |
| Sequencing Platform | Illumina |
Workflow
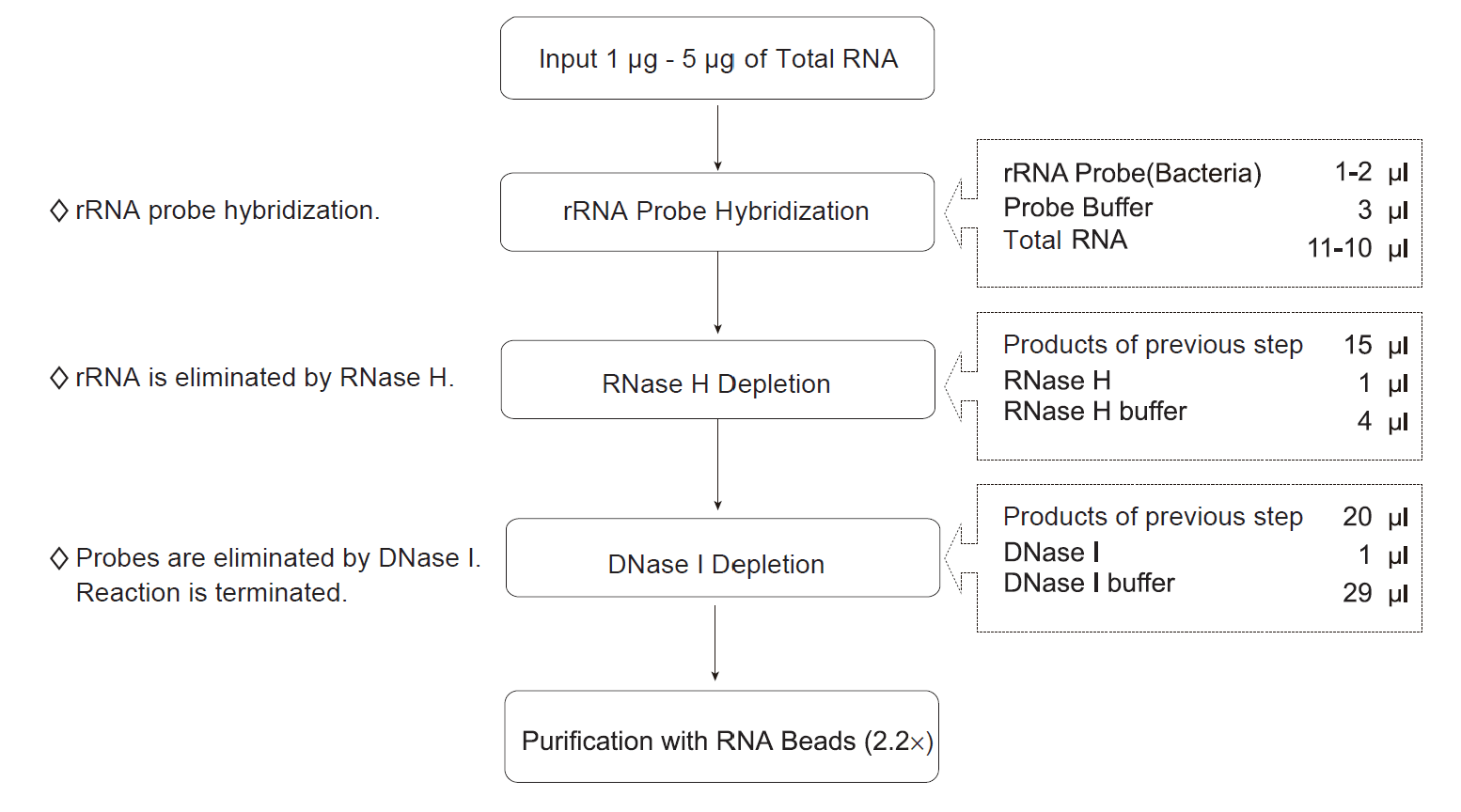 Workflow of CD rRNA Depletion Kit (Bacteria)
Workflow of CD rRNA Depletion Kit (Bacteria)
Protocol
Hybridization of RNA sample and probe
- In a Nuclease-free PCR tube, dilute 1 μg - 5 μg of total RNA in 10 μl or 11 μl of Nuclease-free ddH2O. Then, prepare the reaction solution.
- Put the sample into a PCR instrument and run the program.
Digestion with RNase H
Prepare the digestion with DNase H reaction solution on ice. Mix thoroughly by gently pipetting up and down for 10 times. Collect the liquid to the bottom of the tube by a brief centrifugation. Then put the sample into a PCR instrument and run the program.
Digestion with DNase I
Prepare the digestion with DNase I reaction solution on ice. Mix thoroughly by gently pipetting up and down for 10 times. Collect the liquid to the bottom of the tube by a brief centrifugation. Then put the sample into a PCR instrument and run the program.
Purification of Ribosomal-depleted RNA with CD RNA Clean Beads
- Suspend the CD RNA Clean Beads thoroughly by vortexing, pipet 110 µl (2.2 ×) of beads into the RNA sample of Step 08-3. Mix thoroughly by pipetting up and down for 10 times.
- Incubate the sample on ice for 15 min to make the RNA bind to the beads.
- Put the sample onto a magnetic stand. Wait until the solution clarifies (about 5 min). Then carefully discard the supernatant without disturbing the beads.
- Keep the sample on the magnetic stand, add 200 µl of 80% Ethanol (freshly prepared using Nuclease-free ddH2O) to rinse the beads. Incubate at room temperature for 30 sec and carefully discard the supernatant without disturbing the beads.
- Repeat Step 4.
- Keep the sample on the magnetic stand, open the tube and air-dry the beads for 5 min - 10 min.
- Option A (if the ribosomal-depleted RNA will be used for reverse transcription): take the sample out of magnetic stand, add 20 µl of Nuclease-free ddH2O and mix thoroughly by pipetting for 6 times, and incubate at room temperature without shaking for 2 min. Put the tube back on the magnetic stand and wait until the solution clarifies (about 5 min), carefully transfer 18 µl of the supernatant to a new Nuclease-free PCR tube without disturbing the beads.
- The eluted Ribosomal-depleted RNA is now ready for reverse transcription or RNA library preparation or storage at -20℃.
Option B (if the ribosomal-depleted RNA will be used for RNA library preparation with CD Stranded mRNA-seq Library Prep Kit for Illumina) (RP002): take the sample out of magnetic stand, add 18.5 µl of Frag/Primer Buffer and mix thoroughly by pipetting up and down for 6 times, and incubate at room temperature without shaking for 2 min. Put the tube back on the magnetic stand and wait until the solution clarifies (about 5 min), carefully transfer 16 µl of the supernatant to a new Nuclease-free PCR tube without disturbing the beads for library preparation.



