What is HLA Typing
HLA typing is a laboratory technique used to identify the specific versions (alleles) of HLA genes a person carries. Located on chromosome 6, these genes encode Major Histocompatibility Complex (MHC) proteins, which play a pivotal role in the immune system. By analyzing polymorphisms in these genes, scientists can determine an individual's HLA type. This information is valuable for transplantation research and for studies of disease associations.
HLA Gene Classes Covered by Our Service
Our HLA typing focuses on the classes that encode HLA molecules:
- Class I HLA Genes: This class includes classical loci such as HLA-A, HLA-B, and HLA-C, as well as non-classical loci like HLA-E and HLA-G. The proteins encoded by these genes are expressed on most nucleated cells and are responsible for presenting endogenous antigens. HLA-E and HLA-G, in particular, play important roles in immune tolerance and NK-cell-mediated regulation.
- Class II HLA Genes: This class includes HLA-DR, HLA-DP, and HLA-DQ. These proteins are found on professional antigen-presenting cells and present exogenous antigens.
Note: Class III genes are involved in immune signaling (e.g., complement proteins, cytokines), but do not encode HLA molecules and are not covered in our typing service.
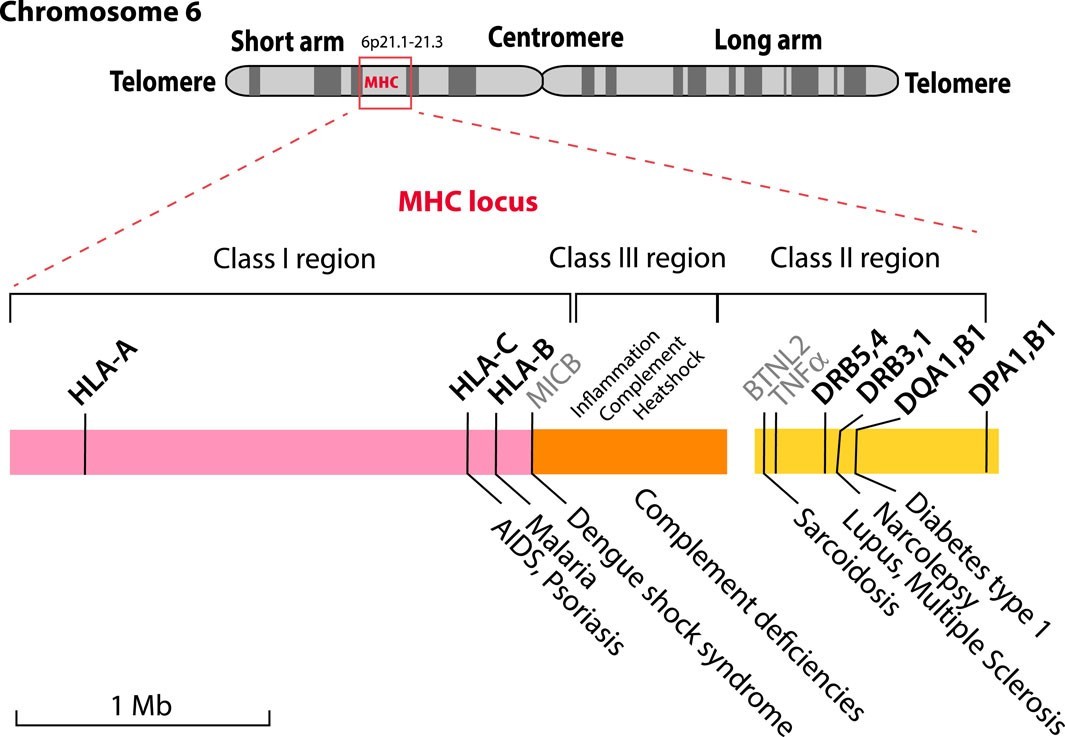 The MHC locus is located on the short arm of human Chromosome 6 with some of the HLA genes indicated. (Wassenaar, Trudy M., et al., 2024)
The MHC locus is located on the short arm of human Chromosome 6 with some of the HLA genes indicated. (Wassenaar, Trudy M., et al., 2024)
HLA Typing Applications in Research
Our high-resolution HLA typing services support a wide range of research areas that rely on understanding immune genetics. Below are some of the key fields where accurate HLA data makes a critical difference:
Population Genetics & Diversity Analysis
Study allele distribution across different populations to uncover patterns in genetic diversity and immune response variability.
Immunogenetics & TCR/BCR Studies
Combine HLA typing with TCR/BCR sequencing to explore antigen recognition and immune system interactions at a deeper level.
Immune Response Profiling
Investigate how HLA variations contribute to individual differences in immune activity, informing biomarker discovery and immune modeling.
Antigen Discovery & Vaccine Development
Utilize HLA data to predict antigen presentation pathways, supporting research in immunogen design and peptide screening.
Reference Panel & Database Construction
Build reliable population reference datasets for downstream association studies or large-scale genomic research.
Choose the Right HLA Typing Method for Your Research
At CD Genomics, we provide multiple HLA typing solutions optimized for different research needs. Whether you're working with large sample sets, targeting rare alleles, or require ultra-high resolution, we offer flexible platforms to support your project.
| Typing Technology | Core Advantages | Supported Loci | Ideal For |
|---|---|---|---|
| Sanger Sequencing (SBT) | - Classic method with clear results - Highly versatile |
HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, HLA-DPB1 (6 loci) | - Projects with defined targets - Moderate sample sizes - Routine research needs |
| Next-Generation Sequencing (NGS) | - High throughput and resolution - Supports automation - Flexible locus selection |
Option for 6 or 11 loci, including HLA-DQA1, HLA-DPA1 | - Large sample volumes - High diversity requirements - Broad locus coverage - Rapid analysis |
| Long-Read Sequencing (PacBio) | - Full-length HLA reads - Eliminates ambiguities - Identifies rare/novel alleles |
Comprehensive coverage of the entire HLA gene range (11 loci and more) | - Research requiring exceptional accuracy - High-resolution typing of complex samples |
| Nanopore Sequencing (NanoTYPE) | - Fast and efficient - Single-tube multiplex PCR - Specialized software support |
HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, HLA-DPB1, HLA-DRB3/4/5 (11 loci) | - Projects needing rapid results - Efficient processing with comprehensive locus coverage |
KIR Typing Service
Building on our high-resolution HLA profiling, CD Genomics now offers precision KIR typing, covering all 14 canonical KIR genes and 2 pseudogenes. Our workflow enables both allele-level resolution and gene copy number variation (CNV) detection, providing comprehensive insights into KIR diversity.
Core Features
- Full gene content & CNV analysis: Detect presence/absence and copy number variation for all KIR loci using quantitative sequencing and read-depth metrics.
- High-resolution allele/haplotype typing: Resolve alleles up to 7-digit resolution using long-read or targeted NGS platforms, enabling confident haplotype inference.
- Seamless integration with HLA typing: Analyze KIR and HLA loci from the same DNA sample using unified laboratory protocols and streamlined bioinformatics pipelines.
Technical Advantages
| Feature | Value |
|---|---|
| Complete KIR panel | 14 loci + 2 pseudogenes, including highly polymorphic and duplicated genes |
| Allele & CNV-level resolution | Up to 7-digit typing via NGS or long-read sequencing |
| Unified workflow | Same input (≥ 1 µg DNA) and pipeline as HLA typing |
| High accuracy | >99% at gene level, >98% allele concordance across validations |
Recommended Applications
- Genetic diversity and population studies analyzing KIR allele frequencies and distributions
- Immunogenetics research investigating KIR–HLA receptor–ligand interactions in disease susceptibility
- Transplantation research modeling donor–recipient compatibility using combined HLA and KIR profiles
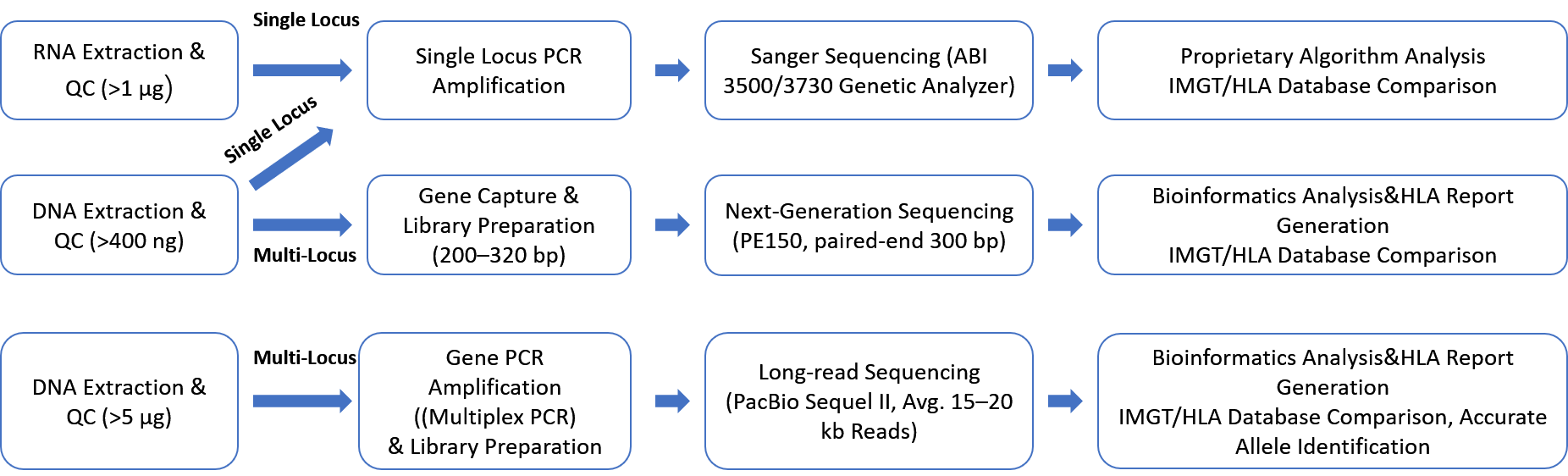
HLA Typing Service Workflow
When precision matters in HLA typing, a reliable and thorough process is essential. At CD Genomics, we’ve developed a meticulous workflow that covers every step—from sample extraction to data analysis—to deliver accurate HLA typing reports tailored to your research needs.
 Note: The IMGT/HLA database we use is the WHO-recommended gold standard for HLA typing.
Note: The IMGT/HLA database we use is the WHO-recommended gold standard for HLA typing.
Detection Range:
| HLA Genotype | Detection Range |
|---|---|
| Single Genotype (Classical Class I and II) | HLA-A, B, C, DRB1, DQB1, DPB1 and non-classical loci HLA-E, HLA-G (Sanger sequencing) |
| 6 Genotypes (Classical Class I and II) | HLA-A, B, C, DRB1, DQB1, DPB1 and non-classical loci HLA-E, HLA-G (Next-generation sequencing) |
| 15 Genotypes | HLA-A, B, C, DRB1, DQA1, DQB1, DPA1, DPB1, DRB3, DRB4, DRB5, DOA, DOB, DMA, DMB (Next-generation sequencing) |
| For additional HLA genotyping needs, please contact the project manager. | |
HLA Typing Bioinformatics Analysis
At CD Genomics, we offer a range of tailored bioinformatics analyses to support your HLA typing needs:
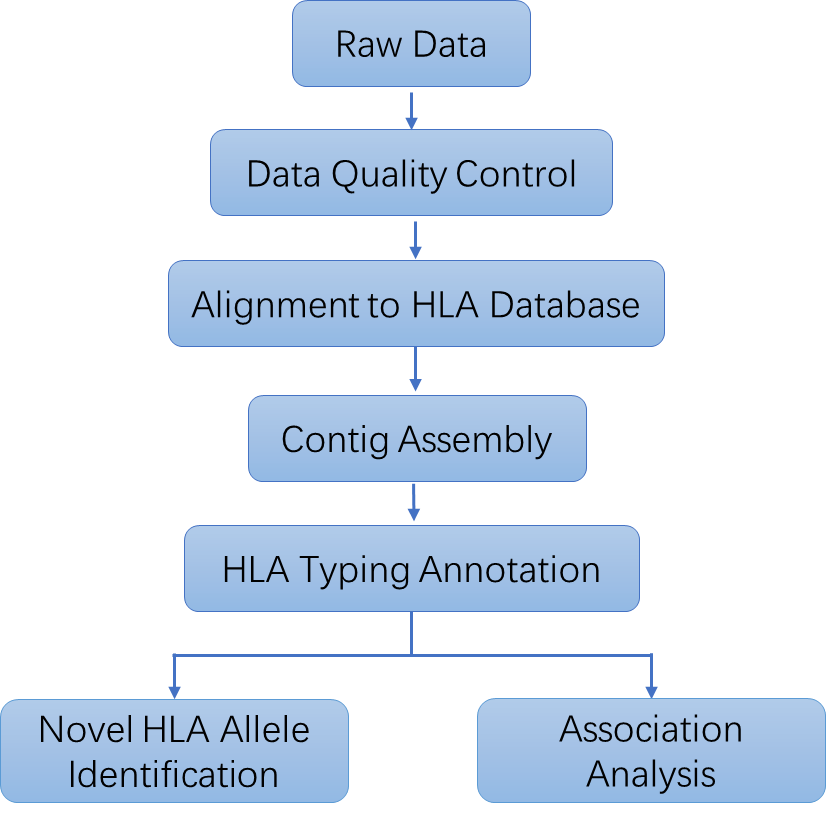
- Raw Data QC:
Thorough quality control of raw sequencing data to ensure accuracy. - Alignment to IMGT/HLA Database:
Precise mapping of sequencing data to the IMGT/HLA reference for accurate allele identification. - Contig Assembly:
Reconstruction of full-length gene sequences for detailed analysis. - HLA Typing and Annotation:
Identification and annotation of HLA alleles for comprehensive results. - Correlation Analysis (Case/Control):
In-depth statistical analysis to explore HLA allele correlations with specific conditions or populations.
Sample Requirements
| Sample Type | Sample Volume |
|---|---|
| Whole Blood | ≥2 mL |
| Peripheral Blood | 3~5 mL; EDTA anticoagulant (purple cap) or sodium citrate anticoagulant (blue cap) |
| Cells | ≥10^6 |
| DNA | ≥1 μg, ≥30 ng/μl |
| RNA | ≥1 μg, >80 ng/μl |
| Frozen Tissue | ≥10 mg |
| FFPE | ≥10 slides |
- Cycle Description: The turnaround time for routine genotyping services varies from several business days to several days, depending on the selected technical platform and sample volume.
Why Partner with CD Genomics for HLA Typing Solutions
In the dynamic world of immunogenetic research, precision and reliability are key. CD Genomics offers top-tier HLA typing solutions, empowering researchers with deep insights. With extensive experience in molecular biology and genomic sequencing, we ensure quality through integrated platforms and efficient operations.
- High Precision and Consistency: Our expertise ensures accurate and reproducible results for your projects.
- Rapid Turnaround: Fast and responsive services keep your research on schedule.
- Validated Data Processes: Rigorous verification at every step guarantees stable data quality.
- Standardized Reporting: User-friendly reports are ready for immediate use in downstream analyses.
At CD Genomics, we’re committed to supporting your breakthroughs in immunogenetic research with dependable and mature services. Let us be your trusted partner in advancing scientific discovery.
HLA Typing: Key Types, Testing Methods, and Transplant Significance
Understanding Your HLA Typing Report
Our HLA typing service provides high-resolution data tailored to support diverse research applications. Here's a quick overview of how to interpret the results.
HLA Typing Resolution Levels
We offer multiple resolution levels depending on the granularity needed:
- Low Resolution – Identifies the broad allele group (e.g., HLA-A02).
- High Resolution – Includes additional specificity at the protein-coding level (e.g., HLA-A02:07).
- Allelic Resolution – Provides full sequence-level detail (e.g., HLA-A01:01:01:01).
- G and P Groups – Group alleles based on shared sequence features in key regions.
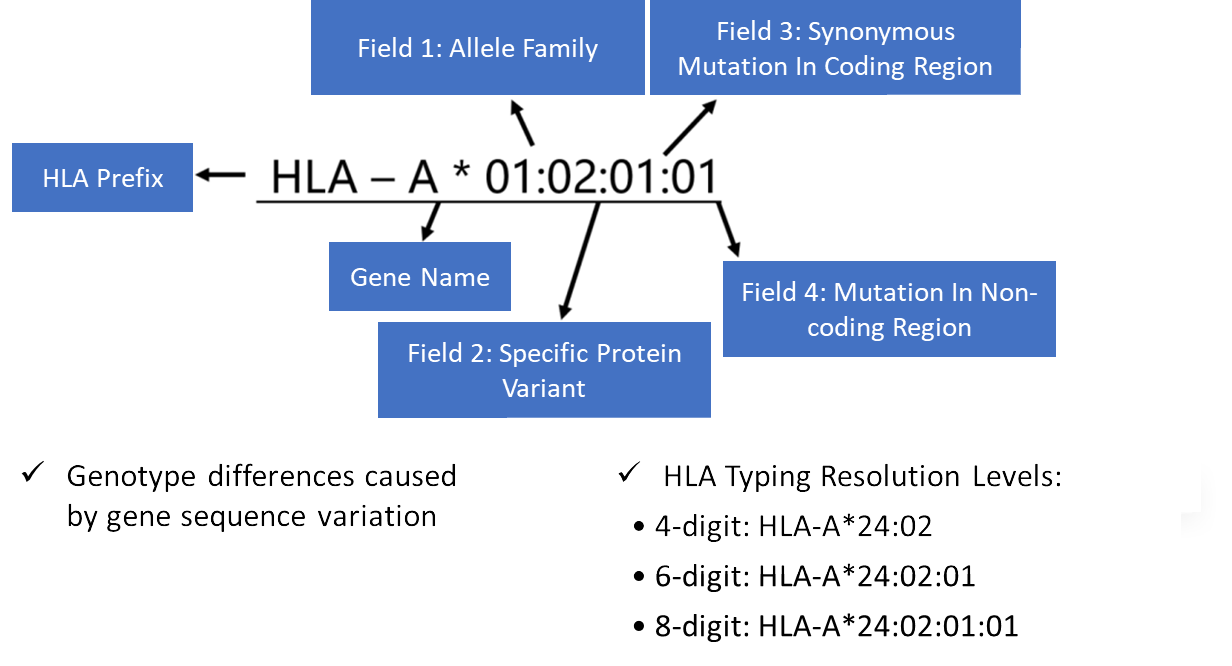
HLA Allele Format – Quick Reference
HLA allele names follow the format: Gene*XX:XX (up to four fields)
- First two digits: Major allele group
- Following digits: Further sequence-level distinctions
Common symbols:
- N – Null (non-functional)
- L – Low expression
- S – Soluble protein
- / or + – Indicates multiple possible alleles
- P / G – Group alleles with identical protein or nucleotide sequences
Reference
- Wassenaar, Trudy M., et al. "DNA structural features and variability of complete MHC locus sequences." Frontiers in Bioinformatics 4 (2024): 1392613. https://doi.org/10.3389/fbinf.2024.1392613
- Mayor, Neema P., et al. "HLA typing for the next generation." PloS one 10.5 (2015): e0127153. https://doi.org/10.1371/journal.pone.0127153
- Sheldon, S., Poulton, K. (2006). HLA Typing and Its Influence on Organ Transplantation. In: Hornick, P., Rose, M. (eds) Transplantation Immunology. Methods In Molecular Biology™, vol 333. Humana Press. https://doi.org/10.1385/1-59745-049-9:157
- Edgerly, C.H., Weimer, E.T. (2018). The Past, Present, and Future of HLA Typing in Transplantation. In: Boegel, S. (eds) HLA Typing. Methods in Molecular Biology, vol 1802. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-8546-3_1
- Wittig, Michael, et al. "Development of a high-resolution NGS-based HLA-typing and analysis pipeline." Nucleic Acids Research 43.11 (2015): e70-e70. https://doi.org/10.1093/nar/gkv184

Table of HLA Type Details (Mayor NP et al., PLoS One. 2015)

Distribution of HLA Allelic Presentation (Nguyen A et al., Journal of virology, 2020)

Distribution of Allelic Presentation for All HLA Alleles and Individually for HLA-A, HLA-B, and HLA-C (Nguyen A et al., Journal of virology, 2020)
References
- Mayor NP, Robinson J, McWhinnie AJM, et al. HLA Typing for the Next Generation. PLoS One. 2015;10(5):e0127153. https://doi.org/10.1371/journal.pone.0127153
- Nguyen A, David JK, Maden SK, et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. Journal of virology. 2020 Jun 16;94(13):10-128. https://doi.org/10.1128/jvi.00510-20
1. What information can be obtained through HLA gene sequencing and information analysis?
Through NGS, HLA typing enables precise identification of specific allele information at each HLA locus, distinguishing between classical and non-classical HLA genes. It also determines specific combinations of HLA allele groups inherited together on a single chromosome. Additionally, NGS-based methods detect previously unreported alleles and ascertain whether each HLA locus is homozygous or heterozygous.
NGS-based HLA typing methods offer advantages of high accuracy, resolution, throughput, and cost-effectiveness. However, the accuracy of NGS-based HLA typing heavily depends on bioinformatics analysis methods, showing significant variability among different approaches.
2. What are the main methods of genotyping?
Serological typing: Focused on the specificity of HLA antigens, this method mainly employs HLA microcytotoxicity tests to analyze HLA types. Serological methods are vital for determining HLA typing and serve as the internationally recognized standard technique.
DNA typing: Concentrating on the analysis of genes themselves, this approach encompasses two main methods: those based on nucleic acid sequence identification and those based on sequence molecular configuration. The nucleic acid sequence identification methods primarily include PCR-RFLP, PCR-SSO, PCR-SSP, PCR-SBT, and the recently emerging next-generation sequencing methods.
3. Is NGS-based HLA typing suitable for clinical applications?
Indeed, NGS-based HLA typing is increasingly being integrated into clinical practice, particularly for organ transplantation, disease association studies, and pharmacogenomics. Its capability to provide high-resolution and precise allele determination enhances compatibility assessments and supports the development of individualized therapeutic strategies.
4. What level of precision is typically required in genotyping?
The specificity of the genotyping should ideally meet the criteria set by the recording of HLA types in the IMGT/HLA database. Differences in the four-digit typing correspond to variations in the encoded proteins, which generally fulfill the demands of scientific research. However, for studies necessitating higher accuracy or specific cases in transplant matching, a six-digit typing may be necessary to acquire more detailed allelic information.
Customer Publication Highlight
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Impact Factor: 8.786 (2022)
Published: 16 August 2023
Background
Adeno-associated viruses (AAVs) are widely used in gene delivery but face challenges due to immune responses against their capsid proteins. CD8+ T cells recognize HLA class I-presented peptides, but the natural repertoire of AAV capsid-derived peptides remains poorly characterized. This study aimed to identify the HLA class I immunopeptidomes of AAV2, AAV6, and AAV9 using mRNA-transfected monocyte-derived dendritic cells (MDDCs) and mass spectrometry. The goal was to map naturally processed peptides, assess cross-reactivity among serotypes, and refine immunogenicity risk assessment for gene delivery systems.
Materials & Methods
Sample Preparation
- 3 healthy donor
- Cell Isolation
- mRNA Transfection
Sequencing
- HLA class I peptides isolation
- HLA typing
- LC-MS data analysis
- Quantification and statistical analysis
- HLA binding prediction
- Conservation analysis
Results
- HLA Class I Immunopeptidome Profiling
- Identified 65 unique AAV capsid peptides (26 from AAV2, 28 from AAV6, 41 from AAV9).
- Peptide Length: 59% 9-mer, 23% 10-13-mer (non-canonical lengths).
- Allele Distribution: 43 peptides bound HLA-B, 15 HLA-A, 7 HLA-C.
Table 1 Donor HLA alleles and frequencies in the US population.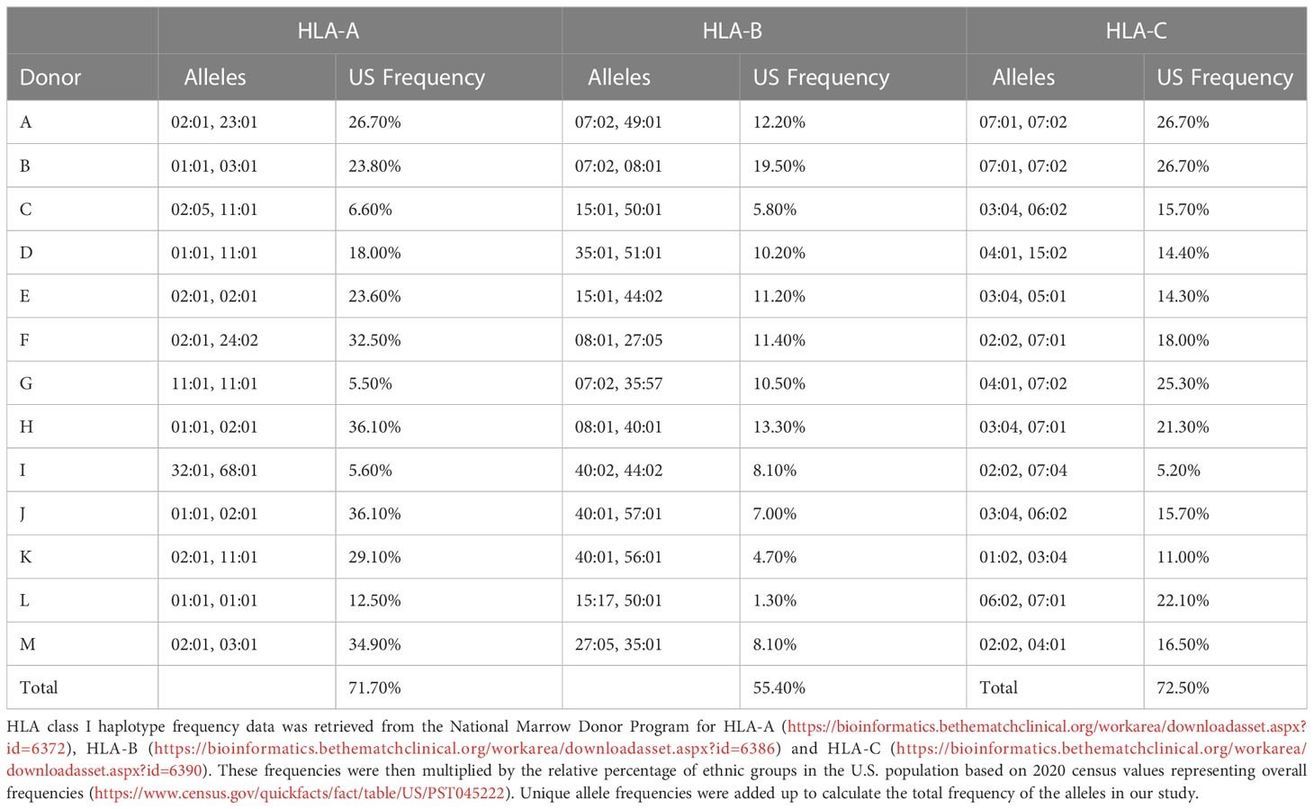
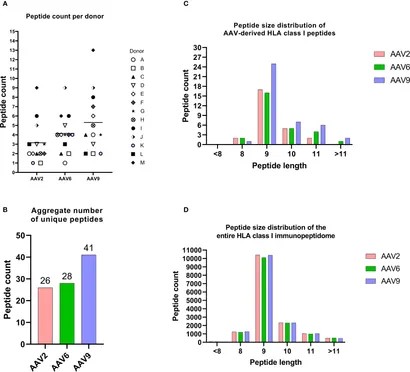 The HLA class I immunopeptidomes of AAV serotypes.
The HLA class I immunopeptidomes of AAV serotypes.
- Comparison with Previous Studies
- Overlap: 6 peptides matched known immunogenic epitopes (e.g., VPQYGYLTL, IPQYGYLTL).
- Novel Peptides: 59 peptides (91% of total) were newly identified.
Table 2 Naturally processed HLA class I peptides and their match with previous identified epitopes.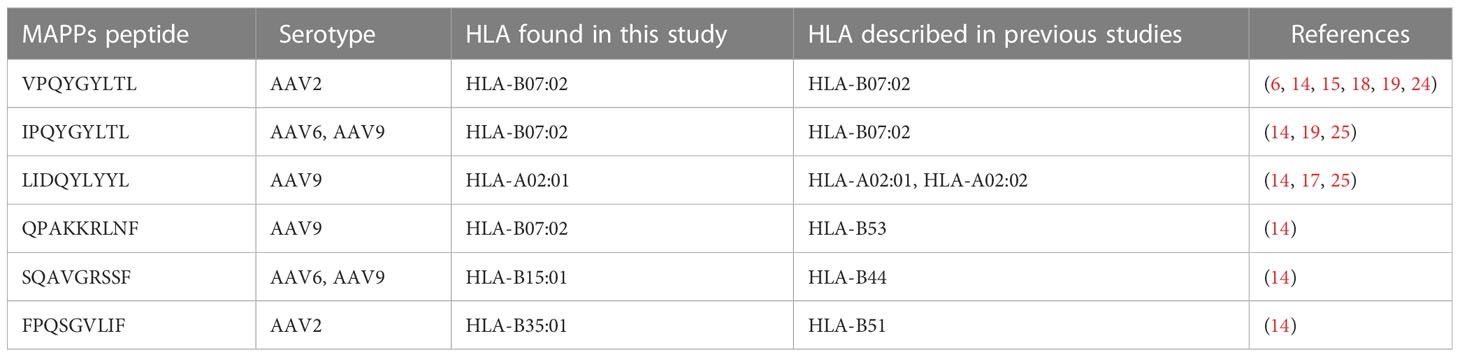
- Conservation and Cross-Reactivity
- 11 Highly Conserved Peptides: Identified across AAV2, AAV6, and AAV9, including regions with single-residue variations.
- Cross-Reactivity Potential: Conserved peptides (e.g., DTSFGGNLGR) may activate CD8+ T cells across serotypes.
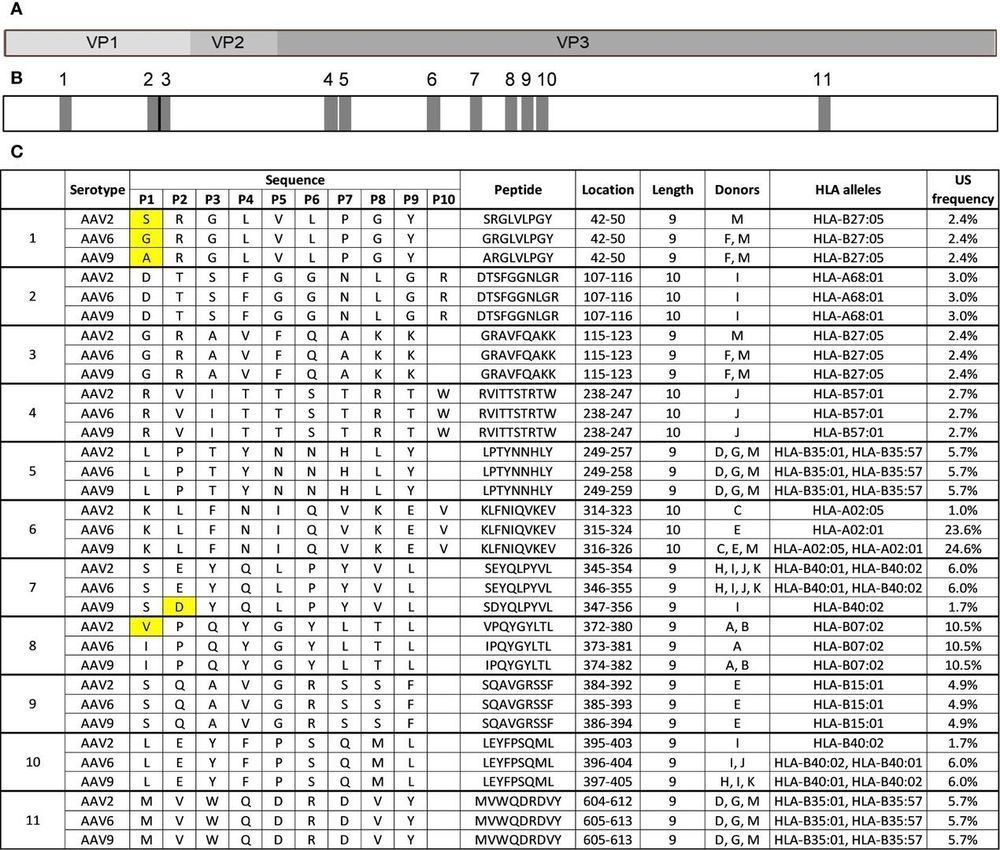 Eleven HLA class I peptides are highly conserved among AAV2, AAV6 and AAV9.
Eleven HLA class I peptides are highly conserved among AAV2, AAV6 and AAV9.
- HLA Class I/II Overlap
- 60% of HLA class I peptides overlapped with HLA class II clusters, suggesting dual presentation potential.
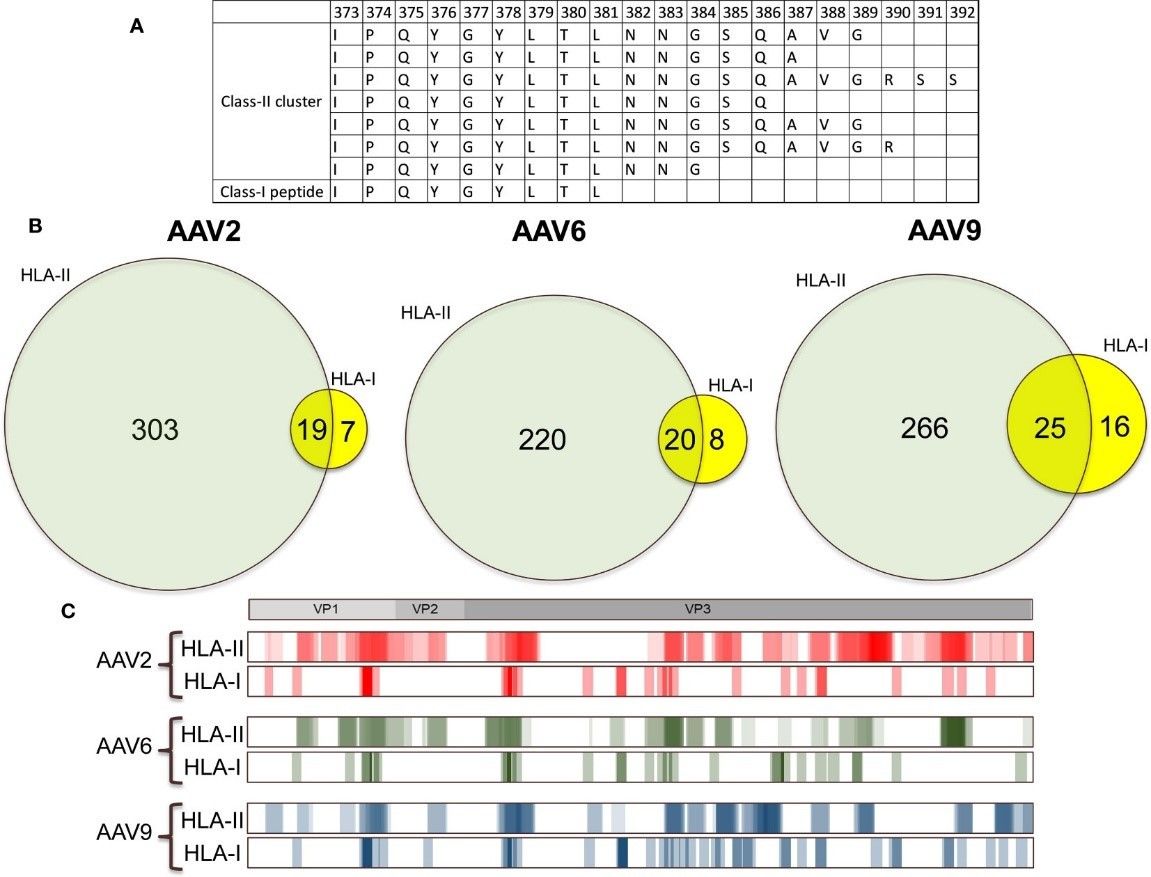 More than 60% of the AAV HLA class I peptides are contained within HLA class II clusters.
More than 60% of the AAV HLA class I peptides are contained within HLA class II clusters.
Conclusion
This study provides the first comprehensive analysis of HLA class I immunopeptidomes for AAV capsids, identifying 65 peptides (59 novel) and highlighting conserved regions with cross-reactivity potential. These findings enhance understanding of AAV-specific CD8+ T cell responses and inform strategies to mitigate immunogenicity in gene delivery systems. Future work should validate the immunogenicity of newly identified peptides and assess cross-presentation mechanisms in experimental models.
Reference
- Brito-Sierra, Carlos A., et al. "The HLA class I immunopeptidomes of AAV capsid proteins." Frontiers in Immunology 14 (2023): 1212136. https://doi.org/10.3389/fimmu.2023.1212136
Here are some publications that have been successfully published using our services or other related services:
The HLA class I immunopeptidomes of AAV capsid proteins
Journal: Frontiers in Immunology
Year: 2023
Isolation and characterization of new human carrier peptides from two important vaccine immunogens
Journal: Vaccine
Year: 2020
Change in Weight, BMI, and Body Composition in a Population-Based Intervention Versus Genetic-Based Intervention: The NOW Trial
Journal: Obesity
Year: 2020
Sarecycline inhibits protein translation in Cutibacterium acnes 70S ribosome using a two-site mechanism
Journal: Nucleic Acids Research
Year: 2023
Identification of a Gut Commensal That Compromises the Blood Pressure-Lowering Effect of Ester Angiotensin-Converting Enzyme Inhibitors
Journal: Hypertension
Year: 2022
A Splice Variant in SLC16A8 Gene Leads to Lactate Transport Deficit in Human iPS Cell-Derived Retinal Pigment Epithelial Cells
Journal: Cells
Year: 2021
See more articles published by our clients.


 Sample Submission Guidelines
Sample Submission Guidelines
