What is Genomic DNA sequencing
Genomic DNA sequencing is a sophisticated method that relies on superior high-throughput sequencing strategies to thoroughly investigate the entirety of an organism's genomic DNA. The core objective of this scientific modus operandi is to attain a profound comprehension of the genetic information encapsulated within the organism. This technique bears exceptional significance in biological research, providing a solid data-based foundation for dissecting the intricate nexus between genes, diseases, and phenotypes. The deployment of genomic DNA sequencing enhances our perception of an organism's biological functionality, the undercurrents driving disease pathogenesis, and the directional progression of biological evolution.
What is the Genomic DNA Sequencing Method?
Currently, DNA sequencing technologies can be succinctly divided into three principal clusters. The initial category encompasses the conventional Sanger Sequencing technology, commonly referred to as first-generation sequencing, extensively recognized as the gold standard in the clinical diagnostic sector. The subsequent category integrates high-throughput sequencing (HTS) or next-generation sequencing (NGS) technologies, both demonstrated to promptly process a massive volume of DNA molecules showcasing unparalleled efficiency. The final category includes single-molecule sequencing technologies, which, devoid of PCR amplification reliance, can directly sequence individual DNA molecules. This latter category is often termed as third-generation sequencing technology.
Sanger sequencing
The Sanger Sequencing Method, an archetypal technique in DNA sequencing, hinges on the close coupling between specific primers and template DNA molecules. Throughout the sequencing process, DNA polymerase catalyzes the progressive addition of the four types of deoxyribonucleotide triphosphates (dNTPs) onto the primer-bonded template DNA. The synthesis of new DNA strands is facilitated by the formation of covalent bonds between the 3' carbon atom of a deoxyribose molecule and the 5' carbon atom of the subsequent nucleotide. This process perpetuates until encountering a terminator, ddNTP, which lacks an oxygen atom at the 3' end, leading to the cessation of DNA strand synthesis.
NGS
Compared to the Sanger sequencing method, high-throughput sequencing technologies such as Illumina sequencing demonstrate superior efficiency, throughput, and cost-effectiveness. Presently, it has emerged as a prevalent method in modern genomics research, finding widespread applications across various fields.
Leveraging NGS technology has successfully facilitated the simultaneous execution of millions of sequencing reactions, marking a notable technical breakthrough. In the past, reliable nucleotide sequences could only be obtained through the coordinated operation of eight different reaction mixtures. Now, however, base sequence information can be directly identified during the synchronous process of sequence extension and detection.
With the advent of NGS technology, the application scope of genomics has significantly expanded. Currently, DNA sequencing has become an integral component in multiple domains, including basic science, translational research, medical diagnosis, and forensic science. Despite notable successes of the NGS technology in cost and time reduction, the relatively short "read length" it generates contributes to demanding computational requirements for subsequent genome assembly. Nonetheless, we remain optimistic that with continuous technological advancement and optimization, these challenges will gradually be resolved.
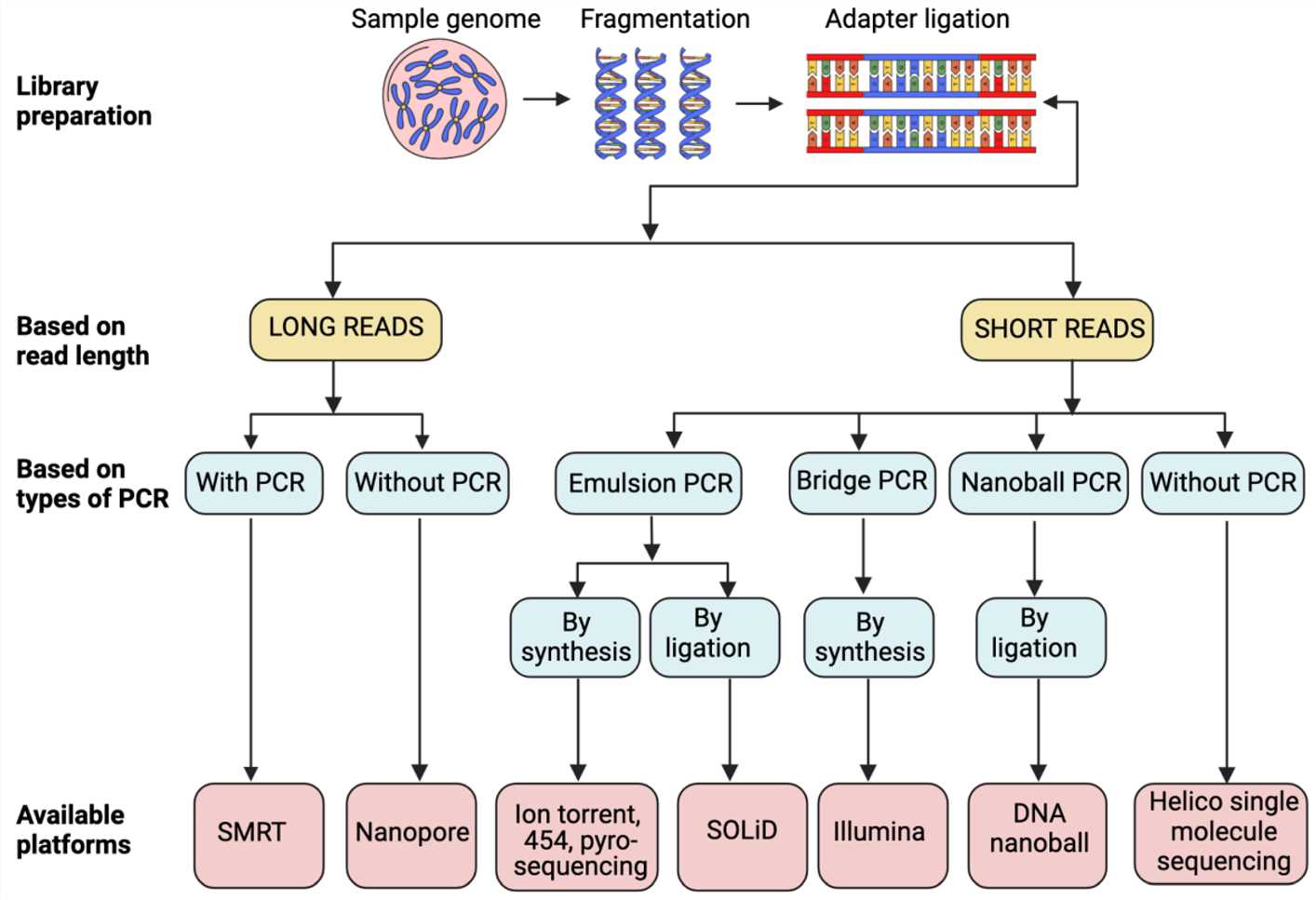 Overview of various NGS technologies (Heena Satam et al., Biology 2023)
Overview of various NGS technologies (Heena Satam et al., Biology 2023)
Single Molecule Sequencing
Single molecule sequencing, also known as long-read sequencing, is gathering increasing attention from the scientific community owing to its superiority in the domain of long sequence reading. Technologies of this kind primarily fall into two categories: Single-Molecule Real-Time (SMRT) sequencing and nanopore sequencing.
Our DNA Sequencing Services
Equipped with advanced NGS platforms, state-of-the-art technologies, and coupled with specialized scientists, CD Genomics delivers a broad array of genomic solutions to meet your diverse research goals and budgets.
-
-
Whole Genome Sequencing (WGS), a revolutionary technique, is now widely used in Human/animal genome research. Its fundamental task is to comprehensively examine and order the complete genome sequence within a biological cell, meticulously capturing all types of mutations from the first to the last DNA. This holds significant implications for deepening our understanding of an organism's genetic information, disease mechanisms, and the relationships between genes and traits.
The development and application of Whole Genome Sequencing extend beyond humankind into other biological sectors. In organisms with an absence of suitable reference genomes or those with low quality reference genomes, de novo sequencing and assembly techniques are markedly valuable. Through WGS, researchers can obtain the complete genome information of an organism, providing important foundations for further investigation into gene functions, genome evolution, gene regulation networks, and more.
In practical applications, Whole Genome Sequencing has achieved globally recognized achievements. For instance, WGS has successfully deciphered the genome sequences of various animals and plants, supplying robust support for research in fields such as agriculture and medicine. Furthermore, WGS is playing a crucial role in pathogenic microbe detection, forensic genetics, and biodiversity studies, among others.
-
-
Within the human genome, the number of exons approximates 180,000, constituting 1-2% of the total genomic entity, around 30MB in computational terms. Pathogenic mutations in the protein-encoding regions of the human genome account for about 85% of the total pathological alterations. Whole Exome Sequencing (WES) is an instrumental technique that privileges the amplification of DNA sequences from exon regions through probe hybridization, prior to undertaking high-throughput sequencing methodologies. The primary objective here is to identify and investigate genetic mutations associated with disease and evolutionary metrics within coding and regulatory regions (Untranslated Regions, UTR). Collating this with publicly-available exome data aids the deeper interpretation of the relationship between various mutations and subsequent diseases mechanisms.
Relative to WGS, Whole Exome Sequencing brings several advantages to the fore: a) Cost-effectiveness: compared with WGS, Whole Exome Sequencing offers superior coverage depth and enhanced data precision, making it an economically preferable choice; b) Depth of sequencing: Sequencing depth can reach an excess of 120x; c) High-throughput capacity: WES is aptly suited for large-scale studies involving numerous target regions; d) High precision: Profound sequencing coverage accompanies high data accuracy, delivering well-optimized outcomes.
-
-
Targeted resequencing is a technique primarily involving multiplex amplicon sequencing and hybrid capture sequencing. It isolates specific genes or genomic regions for sequencing. When compared with WGS and WES, targeted resequencing exhibits the following advantages:
It allows high precision sequencing of vital genes with a deep sequencing exceeding 500x, thereby leading to accurate identification of rare variations.
It is economically efficient, facilitating the study of disease-associated genes.
Capable of identifying variations of allele frequencies as low as 5%.
During a single detection, trustworthy identification of hereditary mutations can be accomplished.
-
-
Mitochondrial DNA (mtDNA) is a molecule within the cytoplasm of cells, where its structure, location, and quantity exert a direct influence on the physiological functionalities and destiny of the living cell. Mitochondrial DNA sequencing technology is a revolutionary biotechnology, which specifically targets and analyzes the sequencing of mtDNA - the pivotal organelle responsible for energy metabolism within the cell.
By revealing an intricate map of the DNA composition within a cell, mitochondrial DNA sequencing technology allows for precise quantification and analysis of DNA structural attributes. This technology paints a detailed portrait of cellular phenotypic characteristics, thereby paving a novel research avenue for scientists. The technology provides researchers with an accurate and rapid means to delve deeply into the cell's inner DNA structure and functionalities. More importantly, it plays a key role in disclosing the mysteries associated with genetic structure of species and their responses to environmental variations.
Hence, mitochondrial DNA sequencing technology does not only furnish researchers with a more efficacious research methodology, but it also serves as a valuable point of reference in the realm of human health and medical research.
-
-
Mitochondria, integral organelle found in eukaryotic cells, bear the responsibility of encoding genes related to their function, actively participating in numerous life processes. Human mitochondrial DNA, characterized by its unique properties and compact double-stranded circular structure, has captivated the scientific community.
Human mitochondrial DNA, approximately 16 kilobase pairs in length, encodes an abundance of genes to support its fundamental role within the cell. Its structural simplicity coupled with the highly conserved coding regions implies an evolutionary stability of its genetic sequence, which greatly aids in functional research.
The maternal inheritance of mitochondrial DNA confers it a unique status in genetic research. As mitochondrial DNA is predominantly passed from mother to offspring, it holds crucial value in the study of inherited diseases. Besides, the rapid evolution rate of mitochondrial DNA underscores its pivotal role in the process of biological evolution.
Simultaneously, the low recombination rate of mitochondrial DNA suggests it holds high value in research on gene mutation and genetic variation. Notably, the high copy number of mitochondrial DNA attests to its vital function within the cell.
To delve deeper into the intricacies of mitochondrial DNA, researchers employ liquid hybridization probe capture technology to enrich mitochondrial DNA, facilitating high-throughput sequencing research. This potent methodology demystifies aspects of mitochondrial DNA and lays a solid foundation for understanding the indispensable role of mitochondria in the theatre of life activities.
-
-
Chloroplasts are among the most crucial and prevalent organelles in plant cells, serving as the central site for photosynthesis. The structure and sequence information of chloroplast genomes hold significant value in revealing the origins, evolutionary changes, and phylogenetic relationships of different species. Simultaneously, chloroplast transformation technology demonstrates considerable potential in genetic improvements and production of bioactive compounds, with the structure and sequence analysis of chloroplast genomes being the cornerstone of this transformation process.
Traditionally, acquiring a plant's chloroplast genome has involved designing degenerate primers using the conservative sequences of the chloroplast genome, amplification of unknown sequences, and long PCR amplification. This amplified product undergoes Sanger sequencing, and the sequences are then assembled to obtain the complete chloroplast genome. However, this process tends to be time-consuming.
With the evolution of scientific technology and emergence of new sequencing tools, numerous researchers have recently shown enthusiasm for high-throughput sequencing. In this approach, chloroplasts are first isolated followed by the extraction of cpDNA. Based on selected references of the chloroplast genome, software is then used to assemble these sequences, ultimately yielding a complete chloroplast genome. Nevertheless, this approach may not be applicable to all species. For example, leaves of higher plants often contain high amounts of pigments and tannins, making isolation of chloroplasts and extraction of cpDNA challenging.
Depending on the specific requirements of the target species, total DNA extraction or cpDNA extraction can be conducted. Information on the taxonomic status is used to search for reference mitochondrial sequences, and degenerate primers are designed for carrying out PCR baiting, unknown sequence amplification, and long PCR amplification. Ultimately, the complete chloroplast genome sequence is acquired through a combination of high-throughput sequencing and Sanger sequencing.
-
-
Plasmids and phages are key tools in modern genetic research, often used for cloning, vector construction, and synthetic biology. Our Complete Plasmid/Phage Sequencing service enables full-length, high-resolution analysis of these genetic elements, helping researchers validate constructs and detect mutations with confidence.
Unlike partial sequencing or restriction mapping, full plasmid/phage sequencing provides complete coverage—including insert, backbone, and regulatory regions—making it ideal for applications such as gene therapy, vaccine development, and microbial engineering. The ability to confirm sequence integrity is especially critical for regulatory submissions and downstream functional studies.
While large or highly repetitive plasmids can be challenging, our platform combines long-read sequencing with advanced error-correction pipelines to ensure accurate assembly and annotation—even in complex constructs.
Why It Matters:
Validates complete plasmid/phage structure, not just inserts
Detects mutations, rearrangements, and off-target modifications
Supports applications in synthetic biology, gene therapy, and vaccine design
-
-
T-cell receptors (TCRs) and B-cell receptors (BCRs) determine how the immune system recognises and responds to threats. Our TCR/BCR-Seq service profiles the full repertoire of these immune receptors, revealing patterns of diversity, clonal expansion, and antigen-specific responses.
By capturing the variable regions of TCR and BCR transcripts, this method enables researchers to track immune dynamics in disease, immunotherapy, and vaccine development. From tumour-infiltrating lymphocytes to autoimmune signatures, the data provide critical insights into immune status and function.
What sets our service apart is the combination of deep sequencing, dual-chain reconstruction (e.g., TCRα/β or IgH/IgL), and tailored bioinformatics that handles hypervariable regions with high sensitivity.
Applications include:
Monitoring T-cell and B-cell clonal expansions in cancer and infection
Characterising immune repertoires in response to immunotherapy
Investigating immune dysregulation in autoimmunity and transplantation
-
-
Amplicon sequencing stands as a potent technique which utilizes specific universal primers to amplify variable regions of the microbes' 16S rDNA/18S rDNA/ITS or functional genes in varied environments. Subsequently, through high-throughput sequencing, we examine the sequence variation and abundance information of the Polymerase Chain Reaction (PCR) products. This approach facilitates the analysis of diversity and distribution patterns of microbial communities within given environments, thereby unraveling the relative abundance and evolutionary relationships amongst the plethora of microorganism species residing within environmental samples.
-
-
Virus Whole Genome Sequencing (VWGS) entails comprehensive genomic sequence analysis of viruses through second and third-generation sequencing platforms. Leveraging bioinformatic methodologies, this approach interprets coding information and makes in-depth inquiries into viral pathogenic systems and the evolutionary trajectory of their genomes. Disciplines across structural genomics and comparative genomics, including differential analysis, homologous gene analysis, collinearity analysis, and species evolution analysis, adopt such techniques to examine these aspects closely. Such efforts improve our understanding of viral diversity, ecology, adaptability, and evolutionary patterns, aiding in predicting the occurrences of emerging infectious diseases.
Research on the chloroplast genome carries significant importance in life science. Its uniqueness is demonstrable in revealing pivotal issues such as species origin and evolution and its applicability extends to other disciplines such as agriculture. With the rapid development of high-throughput sequencing technologies, the study of chloroplasts has become a potent tool probing the origin, structure, and evolutionary issues of cellular organelles. Utilizing second and third-generation sequencing platforms, undertaking high-throughput sequencing on plant chloroplasts, and engaging in-depth sequencing and bioinformatic analysis can yield valuable information about the chloroplast genome sequence, encoding genes, and genetic evolution.
-
-
In the field of genetic research, Long Amplicon Analysis (LAA) has garnered significant attention as a highly efficient and practical method for gene investigation. LAA, a technique grounded in PCR technology, is oriented primarily towards the amplification of targeted gene sequences through the design of specific primers. Unlike traditional PCR amplification, LAA designs primers for the distal portions of the target sequences, enabling the efficient amplification of long DNA fragments. This method boasts high amplification efficiency and accuracy, effectively reducing incorrect amplification due to excessive PCR cycles.
LAA may be deployed to obtain high-coverage genomic DNA fragments, instrumental in unraveling the relationship between genomic structure and function. Through the amplification of specific gene fragments, LAA can proficiently detect gene expression levels across diverse samples, providing a basis for disease diagnosis and treatment. Furthermore, LAA can be utilized for the detection of mutation sites, offering technical support for molecular diagnosis of genetic diseases and tumors, amongst other conditions. Additionally, LAA can serve in the analysis of regulatory elements such as promoters and enhancers, thereby probing the underlying mechanisms of gene expression regulation.
-
-
Shallow Whole Genome Sequencing (sWGS) is among the robust and cost-efficient genomic testing methodologies developed with an aim to expound variations and associated genetic information within an individual's genome. By definition, sWGS signifies partial coverage sequencing of the genomic DNA, with a rather diluted depth and coverage scope compared to deep Whole Genome Sequencing. In practical applications, random fragmenting of the genomic DNA followed by the sequencing of these segments allows a selective access to genomic information. Analyzing these information subsets makes it feasible to detect novel mutations – copy number variations, epigenetic variations, and more.
The advent of high-throughput sequencing platforms like Illumina, PacBio, and Oxford Nanopore has facilitated a rapid evolution of sWGS. Boasting a high sequencing throughput and accuracy, these platforms are capable of sequencing massive quantities of samples in truncated timeframes. Additionally, the progressive refinement of sequencing data analysis tools, including software suites like Bismark and CMap, has substantially optimized the process of sequencing data management and analysis, thereby providing researchers with precise mutation detection results.
In biomedical research, sWGS is invaluable. By comparing the genomes of disease sufferers and healthy controls, meaningful insights into disease-associated gene mutations can be gleaned, thereby providing a sound basis for genetic disease diagnosis and gene therapy. This technique further allows genotypic and phenotypic analyses of organisms, revealing the dynamics between genes and their environment.
-
-
Circulating tumor DNA (ctDNA) refers to fragments of DNA that are released into the blood circulation system from tumor cells. Sequencing these fragments allows us to ascertain genotypic mutation information of the tumor, providing an instrumental basis for tumor research.
The major advantage of ctDNA sequencing resides in its high sensitivity and specificity. Compared to conventional tissue biopsies, ctDNA sequencing can detect tumor mutations at an earlier stage, even when the tumor size is still relatively small. The spectrum of ctDNA sequencing applications is broad, encompassing tumor type identification, molecular subtyping, selection of targeted therapeutic drugs, efficacy monitoring, and prognosis assessment, amongst others.
Nevertheless, ctDNA sequencing confronts several challenges. Firstly, the concentration of ctDNA in the body is relatively low, necessitating detection methods and technologies of high sensitivity. Secondly, the mixture of DNA from normal and tumor cells within ctDNA sequences adds complexity to mutation detection and data analysis. Additionally, interpreting the results of ctDNA sequencing necessitates ongoing research and accumulated experiential insights.
Advantages of Our DNA Sequencing Services
Adherence to the highest standards in providing complete genomics solutions.
Expertise in various aspects of sequencing such as experiment design, target enrichment library construction, and customized bioinformatics analysis.
Delivery of fast, accurate, reliable, and affordable sequencing services.
Capability to provide genomic sequencing services for a wide range of samples including human, mouse, plant, animal and microbe.
Mission to facilitate genomics research by providing access to the latest technologies in the field.
Flexibility to customize services to fit individual project needs and provide specifically tailored analysis.
A consultative approach to identify the best and most economical solutions to meet specific research needs.
For research purposes only, not intended for clinical diagnosis, treatment, or individual health assessments.


 Sample Submission Guidelines
Sample Submission Guidelines
 Overview of various NGS technologies (Heena Satam et al., Biology 2023)
Overview of various NGS technologies (Heena Satam et al., Biology 2023)