The ssDNA-seq Lib Prep Kit for Illumina offers a universal, quick, and easy layout for library preparation in Illumina-based next-generation sequencing (NGS) applications. Damaged, denatured, or otherwise single-stranded DNA, such as gDNA, FFPE DNA, Cell-free DNA, ChIP DNA, viral DNA, ancient DNA, and even single-stranded cDNA, can all be used with the kit. This kit allows library preparation from 10pg to 250ng starting material, making it ideal for NGS library preparation from small sample sizes, damaged, denatured, or ssDNA samples. All Kit elements are subjected to rigorous quality control to ensure that library preparation is consistent and repeatable. This kit is also compatible with a variety of other products.
Species:
Compatible with any species
Storage:
Low-EDTA TE should be stored at room temperature. Other components should be stored at -25℃~-15℃.
Components:
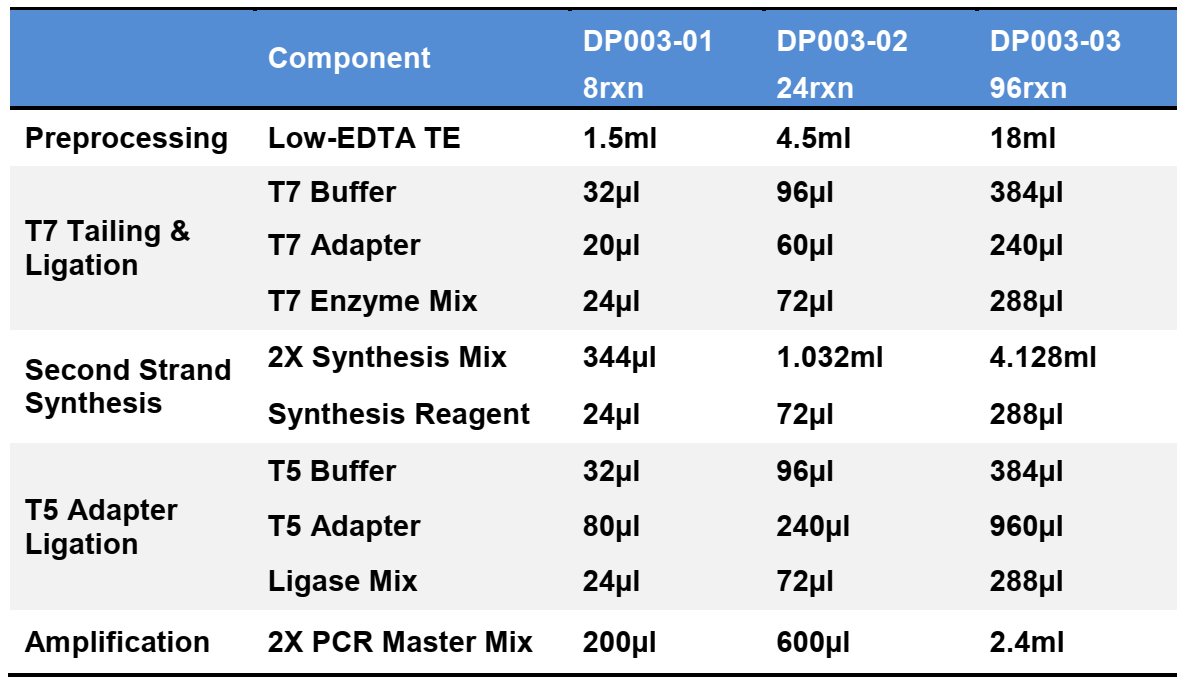
Specifications:
| Sample amount | 10pg-250 ng DNA |
| Features | Fast and convenient workflow enables a single library preparation within 2h. Enables higher library conversion rate, higher and uniform coverage yields. Supports inputs as low as 10 pg. Minimize sequence bias from PCR. |
| Application | It is recommended to use this kit for: Whole genome sequencing ChIP-seq Metagenome sequencing cfDNA-seq Ancient DNA sequencing RNA-seq with first-strand cDNA Hybridization capture sequencing |
| Sample type | Genomic DNA, cell-freeDNA (cfDNA), circulating tumor DNA (ctDNA), formalin-fixed paraffin-embedded DNA (FFPE DNA), Chromatin immuno-precipitation DNA (ChIP DNA), ancient DNA and even single stranded cDNA |
| Sequencing Platform | Illumina |
(Optional) DNA Fragmentation
When working with high molecular weight genomic DNA, the DNA must be fragmented prior to library preparation. If working with samples that have already undergone mechanical or enzymatic fragmentation, such as ChIP DNA, this step may be omitted.
DNA Preprocessing
2.1 Due to the short incubation time of the Denaturation step, pre-assemble all the reagents of the T7 Tailing & Ligation Mix (see next step for recipe), and place on ice.
2.2 Pre-heat the thermocycler to 95 °C.
2.3 Transfer the fragmented DNA sample to a 0.2 mL PCR tube and adjust the volume of the sample to a final volume of 15 µl using Low-EDTA TE, if necessary.
2.4 Place the samples in the thermocycler, programmed at 95 °C for 2 minutes with lid heating ON.
2.5 Upon completion, place tube(s) on ice immediately for 2 minutes. Proceed directly to the T7 Tailing & Ligation step to preserve the maximum amount of ssDNA substrate.
T7 Tailing & Ligation
3.1 Load the Adaptase Thermocycler Program on the thermocycler and pause it at the first step to pre-heat to 37 °C until all samples are loaded.
to pre-heat to 37 °C until all samples are loaded. 3.2 Add 25 µl of the pre-assembled T7 Tailing & Ligation Mix to each PCR tube containing a 15 µl DNA sample and mix by pipetting or gentle vortexing until homogeneous. Spin down.
3.3 Place the samples in the thermocycler and run the program, with lid heating ON.
Second Strand Synthesis
4.1 Load the Extension Thermocycler Program on the thermocycler and pause it at the first step to pre-heat to 98 °C until all samples are loaded.
4.2 Add 46 µl of the already pre-mixed Second Strand Synthesis Reaction (listed in the table below) to each PCR tube containing 40 µl of the T7 Tailing & Ligation mix (step 3.3), using reagents in the order listed.
4.3 Mix by pipetting or gentle vortexing until homogenous. Spin down.
4.4 Place the samples in the thermocycler and run the program, with lid heating ON.
4.5 Ensure the magnetic beads are at room temperature and vortex beads to homogenize the suspension before use.
4.6 Add 105µl magnetic beads (1.2X) to each sample after the Second Strand Synthesis Reaction. Mix by vortexing. Quick spin the samples in a tabletop microcentrifuge.
4.7 Incubate the samples for 5 minutes at room temperature (off the magnet).
4.8 Place the sample on a magnetic rack until the solution clears and a pellet is formed (~ 5minutes).
4.9 Remove and discard the supernatant without disturbing the pellet (less than 5 µl may be left behind). Leave the tubes on the magnet.
4.10 Add 200 µl of freshly prepared 80% ethanol solution to the sample while it is still on the magnetic rack. Use care not to disturb the pellet. Incubate for 30 seconds, and then carefully remove the ethanol solution.
4.11 Repeat step 4.10 once more for a second wash with the 80% ethanol solution.
4.12 Quick spin the samples in a tabletop microcentrifuge and place back on the magnetic rack for 2-3min, remove any residual ethanol solution from the bottom of the tube.
4.13 Add 21µl of Low-EDTA TE buffer and re-suspend the pellet. Mix well by pipetting up and down until homogenous. Incubate at room temperature for 2 minutes off the magnet, then place the tube on the magnetic rack until the solution clears. Transfer 20µl eluate to a new 0.2 mL PCR tube. Ensure that eluate does not contain magnetic beads.
T5 Adapter Ligation
5.1 According to T5 Adapter Ligation Reaction (listed in the support document), add each component In Turn.
5.2 Mix by pipetting or gentle vortexing until homogenous. Spin down.
5.3 Place the samples in the thermocycler programmed at 25 °C for 15 minutes with lid heating ON, followed by a 4 °C hold.
5.4 Ensure the magnetic beads are at room temperature and vortex beads to homogenize the suspension before use.
5.5 Add 40µl magnetic beads (1 X) to each sample after the T5 Adapter Ligation Reaction. Mix by vortexing. Quick spin the samples in a tabletop microcentrifuge.
5.6 Incubate the samples for 5 minutes at room temperature (off the magnet).
5.7 Place the sample on a magnetic rack until the solution clears and a pellet is formed (~5 minutes).
5.8 Remove and discard the supernatant without disturbing the pellet (less than 5 µl may be left behind). Leave the tubes on the magnet.
5.9 Add 200 µl of freshly prepared 80% ethanol solution to the sample while it is still on the magnetic rack. Use care not to disturb the pellet. Incubate for 30 seconds, and then carefully remove the ethanol solution.
5.10 Repeat step 5.9 once more for a second wash with the 80% ethanol solution.
5.11 Quick spin the samples in a tabletop microcentrifuge and place back on the magnetic rack for 2-3min, remove any residual ethanol solution from the bottom of the tube.
5.12 Add 21µl of Low-EDTA TE buffer and re-suspend the pellet. Mix well by pipetting up and down until homogenous. Incubate at room temperature for 2 minutes off the magnet, then place the tube on the magnetic rack until the solution clears. Transfer 20µl eluate to a new 0.2 mL PCR tube. Ensure that eluate does not contain magnetic beads.
PCR Amplification
6.1 According to PCR Amplification Reaction (listed in the support document), add each component.
6.2 Mix by pipetting or gentle vortexing until homogenous. Spin down.
6.3 Place the samples in the thermocycler and run the program, with lid heating ON.
6.4 Ensure the magnetic beads are at room temperature and vortex beads to homogenize the suspension before use.
6.5 Add 50µl magnetic beads (1 X) to each sample after the PCR Amplification Reaction. Mix by vortexing. Quick spin the samples in a tabletop microcentrifuge.
6.6 Incubate the samples for 5 minutes at room temperature (off the magnet).
6.7 Place the sample on a magnetic rack until the solution clears and a pellet is formed (~ 5minutes).
6.8 Remove and discard the supernatant without disturbing the pellet (less than 5 µl may be left behind). Leave the tubes on the magnet.
6.9 Add 200 µl of freshly prepared 80% ethanol solution to the sample while it is still on the magnetic rack. Use care not to disturb the pellet. Incubate for 30 seconds, and then carefully remove the ethanol solution.
6.10 Repeat step 6.9 once more for a second wash with the 80% ethanol solution.
6.11 Quick spin the samples in a tabletop microcentrifuge and place back on the magnetic rack for 2-3min, remove any residual ethanol solution from the bottom of the tube.
6.12 Add 21µl of Low-EDTA TE buffer and re-suspend the pellet. Mix well by pipetting up and down until homogenous. Incubate at room temperature for 2 minutes off the magnet, then place the tube on the magnetic rack until the solution clears. Transfer 20µl eluate to a new 0.2 mL PCR tube. Ensure that eluate does not contain magnetic beads. Stored at -20°C for later use.



