Based on SYBR Green qPCR, the CD Library Quantification Kit for Illumina offers a rapid, sensitive, and precise quantification of library concentration in Illumina next-generation sequencing (NGS). This kit includes all reagents for library quantification, including primers, buffer, enzymes, ROX Reference Dye, DNA standard, and others, in six packages for different uses. The kit is crucial for getting the most out of each sequencing run in terms of data output and quality. Accurate library quantitation is critical for Illumina sequencing to achieve optimal cluster densities, which is required for optimal sequence output.
Application:
This kit is designed for the absolute quantification of the concentration of the Illumina platform's NGS library. Regardless of the way you use for construction, you can use this product for absolute quantification if the end of the library contains Illumina P5 and P7 flow cell binding sequences. The library should be no more than 1 kb in length and no less than 0.0002 pM in concentration. In addition, this product can also be used to detect the library pollution level in experimental environment. Two kinds of primer sequences are provided in the qPCR Primer Mix in this kit:
Primer 1: 5'-AATGATACGGCGACCACCGA-3'
Primer 2: 5'-CAAGCAGAAGACGGCATACGA-3'
Storage:
All components should be stored at -20℃. The Master Mix and ROX should be protected from light. The kits can be valid for 30 freeze-thaw cycles. Once be thawed, the Library Dilution Buffer and the Master Mix can be stored at 4℃ for up to 3 months.
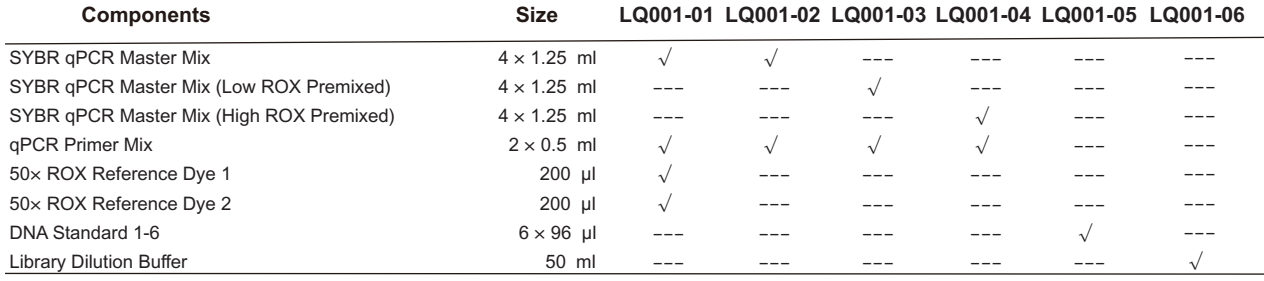
Components:
Specifications:
| Features | Contains all reagents for library quantification. Based on antibody modified hot-start DNA Polymerase, ensures high amplification efficiency, wide GC content adaptability, and high sensitivity. Provides six packages for different uses. Compatible with any mainstream qPCR instrument. |
| Application | This kit is designed for quantification of the concentration of libraries in Illumina next generation sequencing (NGS). The library should be no more than 1 kb in length and must be diluted to the effective mass concentration between 5.5pg/µl to 0.000055pg/µl. Before use, it should be make sure that the preparation library must contain the Illumina P5 and P7 flow cell binding sequences. The qPCR Primer Mix in this kit: Primer 1: 5'-AATGATACGGCGACCACCGA-3' Primer 2: 5'-CAAGCAGAAGACGGCATACGA-3' |
| Sequencing Platform | Illumina |
- Prepare an appropriate volume of library dilution buffer (refers to Notes 2. Library Dilution).
- Dilute the library with Dilution Buffer. The optimum dilution ratio should be adjusted according to the library concentration. The recommended dilution ratio is between 1:1,000 and 1:100,000. At least one additional 2-fold dilution of each library is also recommended (i.e. 1:10,000 and 1:20,000). Library should be kept on ice and diluted freshly before each use.
- Prepare the reaction solution in a qPCR tube according to the handbook.
- Run the program for qPCR.
Note: The Dilution Buffer is stored at 4℃. Equilibrate the dilution buffer to room temperature for 30 min before use and put it back to 4℃ after use.
Notes
- Notes on Library Concentration and Dilution Factor
- Notes on Library Dilution
- Notes on Contaminations and No Template Controls (NTC)
- Notes on Baseline Setting of the Amplification Curve
- Notes on Correction of Library Concentration
- Notes on Melting Curve Analysis
The library must be diluted to the effective Ct range of standard curve for quantitative reaction. Ct values out of the effective range are not suitable for quantification. If more than one dilution of each library is assayed, select Ct values within effective range for concentration calculation.
Dilute the DNA library with an appropriate buffer (Library Dilution Buffer; or self-provide buffer with 10 mM Tris-HCl, pH 8.0 @25 ℃, 0.05% Tween 20). DO NOT dilute the library with water. Use freshly diluted library for quantification. Before qPCR reaction, the diluted library and the thawed DNA Standards should be kept on ice.
A. Inappropriate operation may lead to contaminations in PCR products, which results in inaccurate quantitative results and low credibility. It is recommended to physically isolate the sample preparation area from the template preparation area. Use specialized pipettor and filtered tips. Clean the experimental area regularly (with 0.5% sodium hypochlorite or 10% bleaching agent).
B. Always dispense the DNA Standards from the lowest concentration to the highest (i.e. from DNA Standard 6 to DNA Standard 1). Replace tips after each pipetting.
C. It is highly recommended that no-template controls (NTCs) are included in each assay to detect the PCR specificity and possible contamination introduced. The primers used in this kit is based on universal sequences on Illumina platform. It is normal that amplification product and a Ct value appears in NTC reaction, due to inevitable aerosol pollutions during repeated library dilution. In this case, determine the effective CT range of the standard curve firstly according to the NTC negative control, and then draw the standard curve and calculate the concentration.
The concentration of DNA Standard 1 molar is significantly higher than that of the conventional qPCR templates, therefore generally, its Ct value is as small as 7-9 cycles. While for most qPCR instruments, the default baselines are set at 3-15 cycles, which may increase the Ct value of DNA Standard 1 by mistake and may further affects the linear relationship of the standard curve. Manually set the baseline at 1-3 cycles to avoid this situation.
The fluorescence intensity of SYBR Green, a dye that binds to all double-stranded DNA molecules, is proportional to the molecular weight of DNA molecules. For example, the fluorescence intensity of two 250 bp dsDNA molecules is equivalent to one 500 bp dsDNA molecule. Therefore, it is necessary to correct the library length according to the length of DNA Standards (452 bp) and the average length of the library.
It is recommended to collect melting curve data in every assay, because melt curve analysis is highly important to identify the maximum efficient Ct value and contamination. The melt curves for the DNA Standards displays a characteristic double peak. This is the result of differential local melting in the 452 bp linear template and is not indicative of non-specific amplification. In addition, the molar concentrations of DNA Standards 1-3 are two high, and the amplification products are too many to melt completely at the Tm. Therefore, it is normal that the melting curves of DNA Standards 1-3 sometimes exhibit raising tails.



