Plasmid Isolation Mini Kit is developed specifically for alkaline lysis plasmid purification from 1-5 ml bacterial overnight culture. The Kit offers superior purity plasmid DNA free of proteins and other organic compounds, based on the unique silica membrane innovation and its buffer solution. It has a simple and quick framework, and it can efficiently extract more than 85 percent of bacteria's plasmid DNA.PCR, restriction enzyme digestion, transformation, and sequencing can all be done with the purified plasmid DNA. The Kit can be kept at room temperature (15-25°C) for a long time. Bacterial overnight culture with a volume of 1-5 ml is accepted as a sample type.
Storage:
Stored at room temperature (15-25℃)
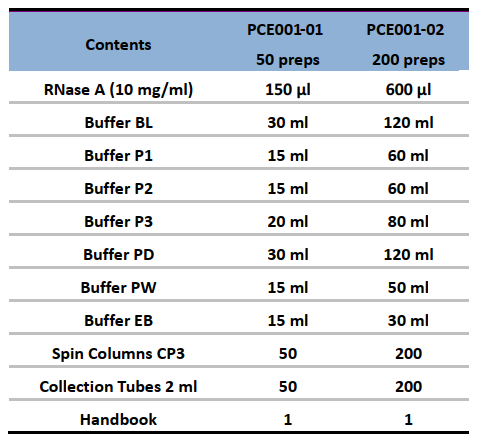
Components:
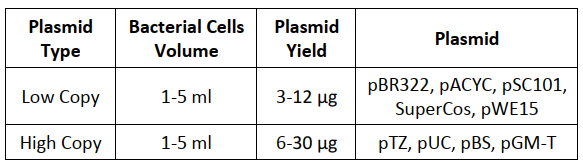
Yield:
Specifications:
| Sample amount | 1-5 ml |
| Features | Fast and convenient workflow. More than 85% plasmid DNA of bacteria can be extracted efficiently. |
| Application | The purified plasmid DNA can be used directly for molecular biological experiments such as PCR, restriction enzyme digestion, transformation, and sequencing. |
| Sample type | bacterial overnight culture |
Add ethanol (96-100%) to Buffer PW before use, check bottle tag for the adding volume.
- Column equilibration: Place a Spin Column CP3 in a clean collection tube, and add 500 μl Buffer BL to CP3. Centrifuge for 1 min at 12,000 rpm (~13,400 × g) in a table-top microcentrifuge. Discard the flow-through, and put the Spin Column CP3 back into the collection tube. (Please use freshly treated spin column).
- Harvest 1-5 ml bacterial cells in a microcentrifuge tube by centrifugation at 12,000 rpm (~13,400 × g) in a conventional, table-top microcentrifuge for 1 min at room temperature (15-25°C), then remove all traces of supernatant by inverting the open centrifuge tube until all medium has been drained (For large volume of bacterial cells, please harvest to one tube by several centrifugation step.)
- Re-suspend the bacterial pellet in 250 μl Buffer P1 (Ensure that RNase A has been added). The bacteria should be resuspended completely by vortex or pipetting up and down until no cell clumps remain.
- Add 250 μl Buffer P2 and mix gently and thoroughly by inverting the tube 6-8 times.
- Add 350 μl Buffer P3 and mix immediately and gently by inverting the tube 6-8 times. The solution should become cloudy. Centrifuge for 10 min at 12,000 rpm (~13,400 × g) in a table-top microcentrifuge.
- Transfer the supernatant from step 5 to the Spin Column CP3 (place CP3 in a collection tube) by decanting or pipetting. Centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through and set the Spin Column CP3 back into the Collection Tube.
- (Optional) Wash the Spin Column CP3 by adding 500 µl Buffer PD and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through and put Spin Column CP3 back to the collection tube.
- Wash the Spin Column CP3 by adding 600 µl Buffer PW (ensure that ethanol (96%-100%) has been added) and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through,
- Repeat Step 8.
- Centrifuge for an additional 2 min at 12,000 rpm (~13,400 × g) to remove residual wash Buffer PW.
- Place the Spin Column CP3 in a clean 1.5 ml microcentrifuge tube. To elute DNA, add 50-100 μl Buffer EB to the center of the Spin Column CP3, incubate for 2 min, and centrifuge for 2 min at 12,000 rpm (~13,400 × g).
Note: No cell clumps should be visible after resuspension of the pellet, otherwise incomplete lysis will lower yield and purity.
Note: Mix gently by inverting the tube. Do not vortex, as this will result in shearing of genomic DNA. If necessary, continue inverting the tube until the solution becomes viscous and slightly clear. Do not allow the lysis reaction to proceed for more than 5 min. If the lysate is still not clear, please reduce bacterial pellet.
Note: To avoid localized precipitation, mix the solution thoroughly, immediately after addition of Buffer P3. If there is still white precipitation in the supernatant, please centrifuge again.
This step is recommended to remove trace nuclease activity when using endA+ strains such as the JM series, HB101 and its derivatives, or any wild-type strain, which have high levels of nuclease activity or high carbohydrate content.
Note: Residual ethanol from Buffer PW may inhibit subsequent enzymatic reactions. We suggest open CP3 lid and stay at room temperature for a while to get rid of residual ethanol.
Note: If the volume of eluted buffer is less than 50 μl, it may affect recovery efficiency. The pH value of eluted buffer will have some influence in eluting; Buffer EB or distilled water (pH 7.0-8.5) is suggested to elute plasmid DNA. For long-term storage of DNA, eluting in Buffer EB and storing at -20°C is recommended, since DNA stored in water is subject to acid hydrolysis. Repeat step 11 to increase plasmid recovery efficiency.



