The Cells and Bacteria Total RNA Kit is specifically designed for total RNA isolation from cultured cells and bacteria samples in a fast, easy, and cost-effective manner, using the unique silica membrane innovation and its buffer solution to eliminate as much protein and genomic DNA contamination as possible. Its key features include (1) optimized buffers for cultured cells and bacteria samples, which allow a fast and convenient process flow for total RNA isolation in just 40 minutes, (2) no phenol-chloroform extraction, which increases workflow safety, and (3) a unique filtration column CS, which eliminates other contaminants. Total RNA that has been purified can be utilized directly in molecular biology experiments such as RT-PCR, Real-time RT-PCR, Northern Blot, and RNA library construction.
Storage:
DnaseⅠ, Buffer RDD and RNase-Free ddH2O (tubular) should be stored at 2-8℃. Buffer RL/β-mercaptoethanol mix can be stored at 4°C for 1 month. Other reagents can be stored at room temperature (15-25℃).
Components:
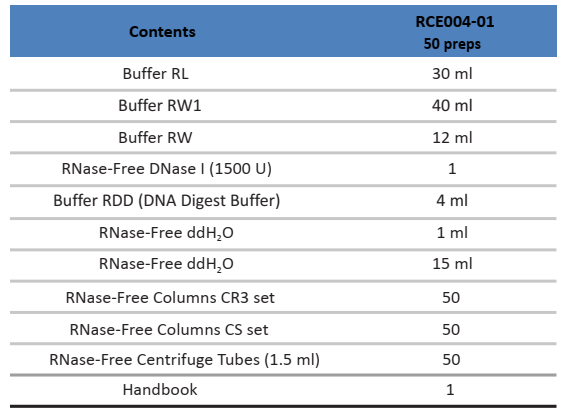
Specifications:
| Features | Optimized buffers for cultured cells and bacteria samples enables a fast and convenient workflow for total RNA isolation within just 40min. No phenol-chloroform extraction enables the workflow more safety. Unique filtration column CS eliminates other contaminations. |
| Application | The purified total RNA can be used directly for molecular biological experiments such as RT-PCR, Real-time RT-PCR, Northern Blot, and RNA library construction, etc. |
| Sample type | cultured cells≤1×107, bacteria samples≤1×109 |
Protocol A: Purification of total RNA from cultured animal cells
- Harvest cells:
a. Cells grown in suspension (do not use more than 1 x 107 cells):
Determine the number of cells. Pellet the appropriate number of cells by centrifuging for 5 min at 300 x g in a centrifuge tube (not supplied). Carefully remove all supernatant by aspiration.
b. Cells grown in a monolayer (do not use more than 1 x 107 cells):
Cells can be either lysed directly in the cell-culture vessel (up to 10 cm diameter) or trypsinized and collected as a cell pellet prior to lyses. Cells grown in cell-culture flasks should always be trypsinized.
(1) To lyse cells directly
Determine the number of cells. Completely aspirate the cell-culture medium, and proceed immediately to step 2.
(2) To trypsinize and collect cells Determine the number of cells. Aspirate the medium, and wash the cells with PBS. Aspirate the PBS, and add 0.10-0.25% trypsin in PBS. After the cells detach from the dish or flask, add medium (containing serum to inactivate the trypsin), transfer the cells to an RNase-Free glass or polypropylene centrifuge tube (not supplied), and centrifuge at 300 × g for 5 min. Aspirate the supernatant completely.
- Disrupt the cells by adding Buffer RL
- Pipet the entire lysate, including any precipitate that may have formed, to an RNase-Free Spin Column CS placed in a 2 ml RNase-Free Collection Tube (supplied). Close the lid gently, and centrifuge for 2 min at 12,000 rpm (~13,400 x g).
- Add 1 volume of 70% ethanol (usually 350 μl or 600 μl) to the cleared lysate, and mix immediately by pipetting. Transfer the sample, including any precipitate that may have formed, to RNase-Free Spin Column CR3 placed in a 2 ml RNase-Free Collection Tube. Close the lid gently, and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through.
- Add 350 μl Buffer RW1 to the RNase-Free Spin Column CR3. Close the lid gently, and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through.
- Preparation of DNase I working solution: Add 10 μl DNase I stock solution (see Preparation of DNase I stock solution) to 70 μl Buffer RDD. Inverting the tube gently to mix.
- Add the DNase I working solution (80 μl) directly to the center of RNase-Free Spin Column CR3, and place on the bench top for 15 min.
- Add 350 μl Buffer RW1 to the RNase-Free Spin Column CR3. Close the lid gently, and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through.
- Add 500 μl Buffer RW to the RNase-Free Spin Column CR3 (Ensure that ethanol has been added to Buffer RW before use). Close the lid gently, and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through.
- Repeat step 9.
- Centrifuge for 2 min at 12,000 rpm (~13,400 × g) to dry the spin column membrane.
- Place the RNase-Free Spin Column CR3 in a new 1.5 ml RNase-Free Collection Tube (supplied). Add 30-100 μl RNase-Free ddH2O directly to the spin column membrane. Close the lid gently, place at room temperature (15-25°C) for 2 min and centrifuge for 2 min at 12,000 rpm (~13,400 × g) to elute the RNA.
For pelleted cells, loosen the cell pellet thoroughly by flicking the tube. Add the appropriate volume of Buffer RL (see the table as below; ensure that β-Mercaptoethanol has been added to Buffer RL before use).
| Number of pelleted cells | Volume of Buffer RL (μl) |
|---|---|
| 5 x 106 | 350 |
| 5 x 106 - 1 x 107 | 600 |
For direct lysis of cells grown in a monolayer, add the appropriate volume of Buffer RL (see the table as below, ensure that β-Mercaptoethanol has been added to Buffer RL before use). Pipet the lysate into a centrifuge tube (not supplied). Vortex or pipet to mix, and ensure that no cell clumps are visible before proceeding to step 3.
Volumes of Buffer RL for Direct Cell Lysis
| Dish diameter (cm) | Volume of Buffer RL (μl) |
|---|---|
| < 6 | 350 |
| 6-10 | 600 |
Protocol B: Purification of total RNA from bacteria
- Collect bacteria by centrifuging for 12,000 rpm (~13,400 × g) at 4°C in a centrifuge tube (do not use more than 1 x 109 cells). Carefully remove all supernatant by aspiration, and proceed to step 2. Other steps should be completed at RT (15-25°C).
- Re-suspend bacteria thoroughly with 100 μl TE buffer containing lysozyme (recipe and incubation time have been showed as follow).
- Add 350 μl Buffer RL (Ensure that β-Mercaptoethanol has been added to Buffer RL before use). Vortex or pipet to mix, and ensure that no cell clumps are visible before proceeding to step 4. If insoluble precipitate has been formed, centrifuge for 2 min at 12,000 rpm (~13,400 × g). Transfer supernatant to another centrifuge tubes.
- Add 250 μl ethanol to the cleared lysate, mix immediately by pipetting. Transfer the sample, including any precipitate that may have formed, to RNase-Free Spin Column CR3 placed in a 2 ml RNase-Free Collection Tube. Close the lid gently, and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through, and put the spin column back to the Collection Tube.
- Add 350 μl Buffer RW1 to the RNase-Free Spin Column CR3. Close the lid gently, and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through.
- Preparation of DNase I working solution: Add 10 μl DNase I stock solution (see Preparation of DNase I stock solution) to 70 μl Buffer RDD. Inverting the tube gently to mix.
- Add the DNase I working solution (80 μl) directly to the center of RNase-Free Spin Column CR3, and place on the bench top (20-30°C) for 15 min.
- Add 350 μl Buffer RW1 to the RNase-Free Spin Column CR3. Close the lid gently, and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through.
- Add 500 μl Buffer RW to RNase-Free Spin Column CR3 (Ensure that ethanol has been added to Buffer RW before use). Close the lid gently, and centrifuge for 30-60 s at 12,000 rpm (~13,400 × g). Discard the flow-through.
- Repeat step 9.
- Centrifuge for 2 min at 12,000 rpm (~13,400 × g) to dry the spin column membrane.
- Place the RNase-Free Spin Column CR3 in a new 1.5 ml RNase-Free Collection Tube (supplied). Add 30-100 μl RNase-Free ddH2O directly to the spin column membrane. Close the lid gently, place at room temperature (15-25°C) for 2 min and then centrifuge for 2 min at 12,000 rpm (~13,400 × g) to elute the RNA.
Note: non-complete removal of culture medium will inhibit lysis and dilute the lysate.
| Concentration of lysozyme in TE buffer | Incubation time (RT) | |
|---|---|---|
| G- bacterium | 400 μg /ml | 3-5 min |
| G+ bacterium | 3 mg/ml | 5-10 min |



