The FFPE Total RNA Kit was developed specifically for isolating total RNA from formalin-fixed, paraffin-embedded tissues and is ideal for a vast range of specimens. Special lysis and incubation situations will cause RNA to be reverse chemically altered by formaldehyde, which will significantly obstruct enzymatic analyses. Furthermore, the lysis buffer releases RNA from tissue sections efficiently while preventing further RNA degradation. Proteins and genomic DNA contamination in FFPE tissues are removed using proprietary silica-membrane technology, a distinctive buffer solution, and a proprietary DNase I. Total RNA that has been purified can be used directly for RT-PCR and Real-time RT-PCR. Store DNase I, Buffer RDD, and RNase-Free ddH2O (tubular) at 2-8°C. Other reagents can be kept at room temperature (15-25°C) for a long time. The sample size required is approximately 2-8 pieces, 5-10m.
Storage:
DNase Ⅰ, Buffer RDD and RNase-Free ddH2O (tubular) should be stored at 2-8℃. Other reagents can be stored at room temperature (15-25℃).
Components:
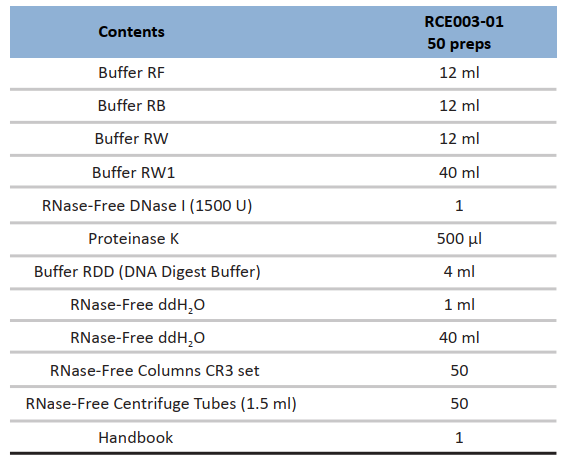
Specifications:
| Sample amount | 2-8pieces, 5-10μm |
| Features | Fast and convenient workflow. Special lysis and incubation conditions reverse chemically modified by formaldehyde of RNA. |
| Application | The purified total RNA can be used directly for RT-PCR and Real-time RT-PCR. |
| Sample type | FFPE samples |
- Using a scalpel to cut the sample block into 5-10 μm thick sections. Note: If the sample surface has been exposed to air, discard the first 2-3 sections.
- Immediately place 2-8 sections in a 1.5 ml RNase-Free microcentrifuge tube, add 1 ml xylene, and vortex vigorously for 10 s.
- Centrifuge for 2 min at 12,000 rpm (~13,400 x g) at room temperature (15-25°C).
- Carefully aspirate the supernatant with pipette, do not disturb the pellet.
- Add 1 ml ethanol (96-100%) to pellet, vortex to mix.
- Centrifuge for 2 min at 12,000 rpm (~13,400 x g) at room temperature (15-25°C).
- Carefully aspirate the supernatant with pipette, do not disturb the pellet (Use a new pipette tip to aspirate the remained ethanol).
- Open the tube lid, incubate for 10 min at room temperature (15-25°C) or 37°C until all residual ethanol has evaporated.
- Add 200 µl Buffer RF and 10 µl Proteinase K into the pellet, vortex to mix thoroughly.
- Incubate at 55°C for 15 min, and then at 80°C for 15 min.
- Centrifuge for 5 min at 12,000 rpm (~13,400 x g) at room temperature (15-25°C), then transfer the supernatant to a new RNase-Free microcentrifuge tube.
- Add 220 µl Buffer RB and vortex to mix.
- Add 660 µl ethanol (96-100%) and vortex to mix (precipitate may forms).
- Transfer 700 µl of solution and precipitate to Spin Column CR3 (put CR3 into collection tube), centrifuge for 1 min at 12,000 rpm (~13,400 x g). Discard the flow-through in the collection tube, and put the spin column back to the collection tube.
- Repeat step 14, until all of the buffer and precipitate go through the Spin Column CR3, discard the flow-through in the collection tube and put the spin column back to the collection tube.
- Preparation of DNase I working solution: transfer 10 µl DNase I stock solution to a new RNase-Free microcentrifuge tube, add 70 µl Buffer RDD, mix gently.
- Add 80 µl DNase I working solution to the center of Spin Column CR3, incubate for 15 min at room temperature.
- Add 500 µl Buffer RW1 to the Spin Column CR3, then centrifuge for 30- 60 sec at 12,000 rpm (~13,400 x g) at room temperature (15-25°C), discard the flow-through and put the spin column back to the collection tube.
- Add 500 µl Buffer RW (Ensure that ethanol (96-100%) has been added) to the Spin Column CR3, incubate for 2 min at room temperature, then centrifuge for 30-60 sec at 12,000 rpm (~13,400 x g), discard the flow-through and put the spin column back to the collection tube.
- Repeat step 19.
- Centrifuge for 2 min at 12,000 rpm (~13,400 x g) at room temperature (15-25°C), discard the flow-through. Open the lid and place the spin column at room temperature for several minutes to dry the membrane completely.
- Place the Spin Column CR3 to a new clean 1.5 ml RNase-Free microcentrifuge tube, and apply 30-100 μl RNase-Free ddH2O to the center of the membrane. Close the lid and incubate at room temperature for 2 min, then centrifuge at 12,000 rpm (~13,400 x g) for 2 min to obtain RNA buffer.



